Instructor Manual
This document describes the experience of the Marseilles University Bioinformatics lecturing team in running the Annotathon. We hope this will help interested readers evaluate how the Annotathon can blend into their own curricula, as well as provide an operational guide, for those who wish to join in. These guidelines are meant to evolve and adapt to your specific environment!
Quick Start Guide
- Find a computer room with web browsers installed and with Internet connection
- Open a new team by emailing the Annotathon list with team name, institutional affiliation and opening/closing dates for the course (can be changed later)
- You will receive your team leader account details (login & password) by email, usually under one working day
- Login to your team leader account, open the "Team" tab and modify team parameters as necessary (opening dates, team code, number of sequences per student cart, sequence origin etc.)
- After opening date, ask your students to create a new account in your team
- Your students can login, add a first sequence to their cart, and start annotating!
- Evaluate annotations that students consider mature (once or twice, your choice)
- After closing date, select the team leader Grades tab to generate an overall grade for each student
- Come back for your next course!
Below is an in depth course management description.
Course Logistics
Students
The Annotathon is run as part of an obligatory course for undergraduate students of biology. The course is run in the third year of preparation for the Bachelor of Science degree (French Licence) with majors in either Molecular Biology or Biochemistry. We have also run the Annotathon with fourth year students preparing a Master of Science degree. Because the course is obligatory, in the core BSc curriculum, cohorts are usually quite large, varying from 30 to 120 students. The course accounts for 3 credits in a total of 30 for that semester (European Credits Transfer System).
Prerequisite: students are required to have taken an introductory Bioinformatics course, in our case an obligatory 30hrs course during their second year. This course introduces resource centers, life science databases and similarity based sequence analysis; a 4x4 hrs hands-on practical in the computer center ensures students have used basic tools at least once in canned exercises (GENBANK, SWISSPROT, PUBMED, dot plots, ORF finder, BLASTp & BLASTx).
Timetable
For each cohort, the Annotathon is run half a day a week, over a relatively compact 8 week period, which starts with three 3 hour lectures covering:
- week 1: Identification of conserved protein domains (patterns, profiles, PROSITE, INTERPRO)
- week 2: Introduction to molecular phylogeny (evolution models, homo-,para-,ortho-logy, tree inference, NJ algorithm)
- week 3: Presentation of the Annotathon: origin of metagenomic sequences, tour of the Annotathon environment, description of annotation fields, practical work organization.
The lectures are immediately followed by four practicals (at a rate of one 4 hours session per week) in the computer center. Because of cohort sizes and limited computing equipment, students are split up into 2 to 4 subgroups and students work in pairs during the practicals. Each student pair has undivided access to a computer terminal during the 16hrs of supervised practicals.
We start the first practical session by having student pairs create a new Annotathon account in the relevant team, and then spend half an hour reading the Rule Book. The rest of the first session is then devoted to explaining the analysis workflow step by step (going through each editable field of the sequence annotation form), with regular assemblies around the videoprojector screen for general briefings and demos. The last three practicals are unstructured and entirely devoted to helping out individual students with their specific analyses results and interpretations. We have also found it useful to start the last three sessions with a 20 minute review of interesting annotations issues faced by students (videoprojection of annotations).
Each student pair is assigned three metagenomic sequences to annotate over the course of the 4 week practical. It is made very clear to students that to complete their annotation assignments, they will be required to work outside supervised practical sessions. Two half days of homework is a suggested minimum, with Annotathon computer logs showing that students are connected on average for 46 hrs (min=16hrs, max=109hrs), representing an average of 30 hrs homework over the 4 week period.
The computer center is thus in free access 2 hours every lunch time and two hours every evening (longer evening access would be much preferable). We have found that with the generalization of home broadband Internet connection, students tend to prefer working on private computer equipment.
An Annotathon closing date and time is agreed on at the beginning of the course, usually around 10 days after the last practical session. Annotations can no longer be edited after the closing date.
Equipment
Other than access to an amphitheater for the 10hrs of lectures (preferably with Internet access and video projector), the course requires access to a networked computer room with one terminal per student (or pair of students) for the supervised practicals, as well as ample computer center free access outside teaching hours. A terminal should also be available for one of the instructors for live team management tasks (see Team Management) and demos (see video projection below).
A video projector connected to the instructor's terminal is most useful in the computer center during the first practical session, preferably with a switch allowing either ordinary monitor display or video projection.
The nature of the computer center hardware or operational system (OS) is irrelevant, as long as the terminals have an Internet browser with cookies and javascript activated (Firefox, Opera, Safari and Internet Explorer browsers have all been used successfully). We usually run the practicals in a four room computer center, equipped with a total of 80 UNIX terminals running the Gnome user interface.
Instructors
Our experience suggests that a minimum of one instructor per 20 students is necessary for a successful Annotathon based course. Instructors not only supervise the practical sessions, but are also each assigned an equal quota of the student annotations to evaluate over the 5 weeks duration of the Annotathon (see Evaluation below). Instructors are university Bioinformatics professors and assistant-professors. We also seek the assistance of postgraduate students (usually bioinformatics PhD students) to help with the practical sessions.
We have found that adequate instructor to student ratios are of paramount importance during the practical sessions, especially the first and second sessions, where students need to be made rapidly confident with the interface and analysis workflow. If the student direct questioning intensity returns to more reasonable levels after that, the online evaluation of student annotations, which picks up at that point, is also highly demanding for instructors. According to Annotathon connection logs, instructors spend on average 30 hrs evaluating student annotations (20 students pairs per instructor, three sequences evaluated twice per student pair, see Evaluation below). The team leader instructor is usually connected a further 20 hrs with account management, forum questions etc.
Evaluations
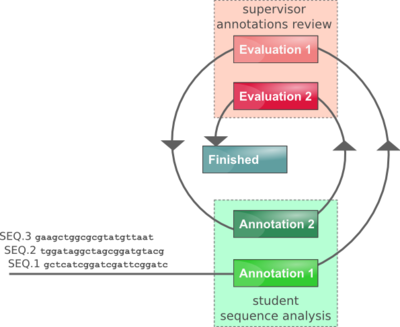
We recommend that each student annotation is evaluated twice (allowing students to improve their annotations following initial instructor comments), although the Annotathon can also be configured for single pass annotations/evaluations (see below #Annotathon Team Management). In the case of two stage evaluation, the procedure goes as follows:
- student adds sequence to his cart (editing mode enabled)
- student annotates his sequence until he is satisfied it is ready for initial review
- student submits his annotated sequence for evaluation 1 (editing mode temporarily disabled)
- instructor receives email notice that evaluation 1 is pending
Note: while waiting for the evaluation, student can add a new sequence to his cart
- instructor evaluates the first pass annotations (assigns a mark and provides comments, see below)
- student is notified that initial evaluation is ready (editing mode enabled)
- student amends his annotations until he is satisfied they are ready for final review
- student submits his annotated sequence for evaluation 2 (editing mode permanently disabled)
- instructor receives email notice that evaluation 2 is pending
- instructor evaluates the second pass annotations (assigns a mark and provides comments, see below)
- student is notified that final evaluation is ready (annotations and comments available in read only mode)
Each instructor evaluation, of each annotated sequence, consists of two parts:
- instructor free text comments
- a numerical mark between 1 and 10
In the case of three sequence assignments per student combined with iterative double evaluations, each student will receive 6 numerical marks by the end of the course. After Annotathon closure, numerical marks are first normalized across instructors (median subtracted and divided by standard deviation). The final Annotathon grade for each student is then computed as the average of the 6 instructor normalized individual evaluations.
In addition, instructors can grade the intrinsic difficulty of each sequence on a 1 very easy to 5 very difficult scale. These difficulty factors are used to give more weight in the final student grade to annotations that required extensive work, e.g. ORF's with numerous homologs presenting complex phylogenetic relationships are more difficult and represent more work than a non coding sequence with no detectable homologs. Individual evaluations are simply repeated difficulty factor times in the computation of the evaluation mean, used to derive each student grade.
It is important that students submit their annotations for evaluation regularly, rather than wait until the deadline. One measure that helps enforce regular submissions is to only allow new sequences to be added to the student cart, after the preceding sequence annotations have been submitted for evaluation. This ensures that students get evaluation feedback early on in the practicals, allowing them to take comments into account with subsequent sequences. This also helps instructors spread out their evaluation duties.
With some teams we also added an extra session where each student pair presented one of their three annotated sequences to the class, with the help of a video projector. The 5 minute presentation would be followed by 5 minutes questions by instructors or fellow classmates. The presentation would give rise to an additional individual mark for each student.
Because our students work in pairs during practicals, the course overall grade is composed for 50% by the Annotathon grade generated for the pair, and for the other 50% by a individual written examination for each student.
Annotathon Team Management
Annotators, instructors & team leaders
An Annotathon team consists of:
- annotators (a student cohort)
- instructors (evaluate the student annotations)
- a team leader (instructor who manages the team)
Students, instructors and team leader each have individual Annotathon accounts with specific roles and privileges. Students annotators add sequences to their sequence cart for subsequent annotation. Instructors answer student questions and evaluate their annotations (instructors are each assigned an equal quota of the annotations to evaluate). As well as being an instructor (therefore takes part in evaluations), the team leader has special team management privileges (such as changing the team's configuration parameters, inviting new instructors into the team, or responsibility for producing the final grades).
Creating a new team
To create a new team for you and your students, please email the Annotathon list, providing the following information required for team creation:
- team name (20 characters, will appear on the Annotathon website)
- name of host institution (e.g. university or college name) and city
- team leader preferred username
- team leader email address
- team secret code (share this with your students, needed for them to join team)
The team leader will receive account details (including password) by email. We normally open new teams by the next day.
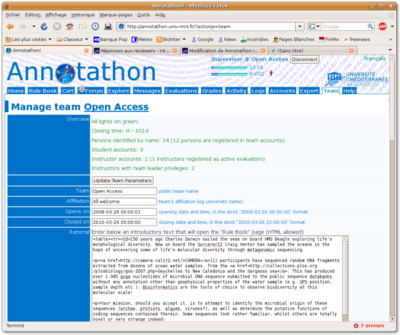
General team configuration
Log into your team leader account and click on the Team tab. Your team will be initially configured in the same way as the Open Access generic team. At the top of the Team tab, a short summary recalls essential data about the team (number of registered students, number of opened accounts, number of instructors etc.)
Review and amend, as necessary, the following parameters of the Team tab form:
- Public team name
- Official affiliation (e.g. university or college name)
- Team code (a short code required by students to be allowed to create an account in your team)
- Start date for student annotations
- Closing date for student annotations
- Team code (this is used to restrict new account creations in your team to your students)
- Three introductory texts that will appear in the student Rule Book (Rational, Team information & Evaluation)
Remember to click on the Update Team Parameters to save your modifications.
Interface language
The default language used for the Annotathon interface (messages, buttons etc.) can be selected using the Team->Language parameter. Users can always select which interface language they personally use, by clicking on the language links in the top hand corner of the Annotathon pages. Currently supported languages are English and French (offers to translate the interface in further languages are warmly welcomed, see about page)!
Inviting new instructors
Under Team->Instructors are listed the instructors currently associated with the team. To invite a new instructor, enter his username and email in the fields provided. He will automatically receive his login parameters by email.
Instructors whose usernames are ticked are considered active evaluators, i.e. will be assigned their share of annotations to evaluate as students add new sequences to their carts. Deselect an instructor so that he receives no additional sequences to evaluate (annotations assigned to him previously remain his responsibility).
Student identification
Enter in the Team->code box a short text (e.g. "RNA" or "firmicute") that will be required by students when they first create their account. This is to help reduce ghost visitors joining your team, or your students joining the wrong team by accident.
Enable the Team->Predefined list of student names in order to make students select their names in a predefined list of names (after saving this modification, you will then need to upload a CSV formatted file of student names). If left disabled, students are allowed to enter their names, as free text, on creating their account. Encourage students to enter their email addresses at account creation (they can enter several addresses separated by commas). This allows them to receive in real time through their email inbox "evaluation ready" notices, team leader announcements etc. If entered, the email address will need to be validated, by clicking on a special link in an email delivered to that address. If the email is never received, students can log in and correct their email addresses using their Profile tab.
The Team->Number of student work groups is only used if a large student cohort is split into subgroups, during practicals. This is informational unless you choose the RECYCLE sequence dispatch scheme (see below).
To prevent brute force account hijacking, accounts are frozen after 6 authentication failures (bad login/password combination). See below #Frozen account unlocking for thawing instructions.
Student account management
Modifying accounts
Open the instructor only Accounts tab and select which account you wish to modify (select account either by login name, email or student names). Whereas the student Profile tab only allows editing of password and email address, the instructor Accounts tab allows editing of all account fields (login name, password, student names, subgroup and email addresses). To help trace problems, Annotathon log entries, relating to the corresponding account, are also displayed at the bottom of the Accounts tab.
Frozen account unlocking
To avoid account hijacking, user accounts are frozen after 6 authentication failures. The users then receive an email containing a special link which, when followed, thaws the user's account. In case students entered an erroneous email address, frozen accounts can also be unlocked from the instructor only Accounts tab: simply select the relevant account and press Reset login failures.
Monitoring activity
The instructor only Activity tab displays a general information sheet on the team's activity. For each team account are indicated number of sequences in cart, connection state (logged in/out), last IP address, etc.
Full Annotathon logs
The instructor only Logs tab displays a anti-chronological list of the Annotathon system logs. The logs are usually only useful when trying to pin down issues during the Annotathon (such as dating or verifying the authenticity of certain actions). Use the << or >> buttons to browse through earlier or later logs.
Sequence dataset
Selecting sequence source
Select the Team->Sequence data set from which students will be allowed to choose sequences, when adding sequences to their carts. The Global Ocean Sampling expedition dataset is selected by default. The GOS dataset having not been uploaded in full, click on the Sequence reserve link to see how many sequences have been uploaded and how many are still available (i.e. not annotated yet). If the dataset selected for the team has several defined samples (see below), then students will be asked to select from which sample location they wish to pick a new sequence, when adding new items to their sequence cart.
Updating & adding new sequences to a dataset
Before modifying a dataset, please contact the webmaster in order to make sure dataset integrity is maintained (for instance we need to make sure existing sequences are not redundantly duplicated). If you wish to use non oceanic metagenomic sequences, please follow the #Creating a new dataset section, to create a new independent dataset of your own.
If you wish to upload more sequences to an existing dataset or create an entirely new dataset, click on the To add more sequences to the database... link. Create a tab delimited text file with one sequence per line as shown below. If necessary, this file can also contain sample definitions (lines starting with #SAMPLE). The format is:
#SAMPLE DATASET_NAME SAMPLE_ID SAMPLE_LOCATION COUNTRY DATE TIME GPS DEPTH TEMP SALINITY PORE_SIZE HABITAT GEO_LOC SAMPLE_ID SEQ_ACC_NUM SEQ_START SEQ_END DNA_SEQUENCE
A valid dataset file defining two samples and 3 sequences could look like this:
#SAMPLE Global Ocean Sampling expedition JCVI_SMPL_1103283000001 Sargasso Sea, Station 11 Bermuda (UK) 02/26/03 10:10:00 31°10'30n; 64°19'27.6w 5 20.5 36.7 0.1-0.8 Open Ocean Sargasso Sea #SAMPLE Global Ocean Sampling expedition JCVI_SMPL_1103283000021 Off Key West, FL USA 01/08/04 06:25:00 24°29'18n; 83°4'12w 1.7 25 36 0.1-0.8 Coastal Caribbean Sea JCVI_SMPL_1103283000001 JCVI_READ_1098127013050 1 938 TGCTCGCCGCGCTCTGGGGTTCA[...snip...]ACCCTGATGGACTTAGCCCTTGGCTGGACTAC JCVI_SMPL_1103283000021 JCVI_READ_1098127013057 1 927 TGGACCTCTTCCAATGTCTGCTC[...snip...]CCATCCGGTCTAAATGTACAAGTTTCCACT JCVI_SMPL_1103283000021 JCVI_READ_1098127013056 1 942 CCAATGTCTGCTCCACCACCTTT[...snip...]TTTGCTACTTGCACCCATATCTCACCTTGATAACCT
Make sure you respect the exact GPS syntax (31°10'30n; 64°19'27.6w) for correct sample map positioning. If you specify existing SAMPLE_ID's or existing SEQ_ACC_NUM's
(which you created), then the corresponding samples or sequences will
be updated accordingly. If non existing id's are specified, then new
items are created.
Creating a new dataset
To create a new dataset, just specify a new DATASET_NAME that isn't present in the database (use the Sequence reserve link to list available dataset and sample names).
Student sequence carts
The Team->Cart sequence dispatch parameter allows you to switch between NOVEL and RECYCLE modes. The default NOVEL mode ensures that only new sequences (never before annotated by any Annotathon team) will be added to student carts. In RECYCLE mode (deprecated), sequences are only ever dispatched once per team (so that a given sequence is only annotated by one student pair, inside a given team), however the same sequences are redundantly dispatched across separate teams (this allows instructors to compare parallel annotation during evaluations). If your team is split into subgroups, then, in RECYCLE mode, sequences are uniquely dispatched inside a subgroup and redundantly dispatched across distinct subgroups.
A third CONVERGENT sequence dispatch mode is still at development stage: this mode will be identical to the RECYCLE mode, however sequences will only be recycled through separate teams, until the main controlled vocabulary annotations converge. Convergence is reached when GO Biological Process, GO Molecular Function and Taxonomic Classification are identically assigned to the same sequence, independently, by three separate teams.
Specify in the Team->Maximum number of sequences per cart field how many sequences each annotator account will be expected to annotate in total. We have found three sequences is reasonable for students following a 30hr course representing 10% of semester course work. Significantly higher number of sequences results in lower quality annotations, whilst less than three sequences provides less opportunity for students to experience a variety of different sequence flavors.
Finally use the Team->Maximum number of sequences in Annotation 1 phase field to enforce regular submission of annotations for evaluation. When set to 1 (recommended), students can not add a new sequence to their cart, until all previous sequences have been submitted for evaluation.
Sequence identifiers
On top of the SEQ_ACC_NUM associated with each sequence (provided by the dataset designer, preferably an accession number from a public database), note that each sequence is assigned two additional types of identifiers:
- the internal database identifier (a simple integer) assigned by the Annotathon at dataset upload time and which is unique, within the scope of the database. This private identifier is only ever visible in the first column of the Evaluations tab (see #Instructor evaluation tools)
- the Annotation code which is the code associated with student's annotations of a specific sequence (the only identifier students see). This code is in the form GOS_AB123450, where the first three letters identify the sequence dataset, the following two letters are random, and the following number is an Annotathon wide integer, incremented by ten every time a sequence is added to a student cart. This Annotation code is used throughout the Annotathon to identify student annotations.
If the same sequence is redundantly annotated by three distinct students (in RECYCLE or CONVERGENT dispatch modes), there will be three distinct Annotation codes attached to this specific sequence.
Sequence versioning & reverting to a previous version
A annotation versioning system is in operation, with the number of the latest version indicated after a dot at the end of the Annotation code, e.g. GOS_AB123450.56. Previous versions of student annotations can be viewed, or annotations can even be reverted back to a previous version (last 100 versions are stored, older versions are deleted). Both students and instructors have access to this functionality.
To view previous annotation versions, simply select the required version in the popup list at the top of each annotation page (either from the cart View mode, from links in the Explore tab or from View links in the instructor Evaluations tab). Once the previous version is displayed, it can be reverted to by pressing the Replace GOS_AB123450 with version 42 button.
Evaluations
Evaluation scheme
The Team->Evaluation schema parameter controls the number of times student annotations are evaluated by instructors. We recommend the default DOUBLE setting, where we allow student to respond to instructor's comments and criticisms during the Annotation 2 phase. This greatly accelerates training and produces final annotations of a far superior quality. Collaterally, it must be stressed that this naturally increases the instructor workload, by doubling the number of evaluations. However, we have found second evaluations much easier to produce, since the sequence context is familiar from the first evaluation, and annotations require less commenting, after students have taken first round comments into account. Nonetheless, Team->Evaluation schema can be set to SINGLE to disable second stage annotations and evaluations.
You can use the Team->Number of best evaluations to consider for grade parameter to take only a subset of the evaluations into account, during the final grade computation (see below Computing overall grade). For instance, if you have set the Team->Maximum number of sequences per cart to 3, and you set Team->Number of best evaluations to consider for grade to 2, then only the two best annotations, for each student, will be used to compute the final grade (i.e. the worst annotation will be ignored). If set to zero (default), all available annotations will contribute to final grade.
The instructor evaluation form has both free text comments fields and predefined generic comments (for those frequently occurring mistakes). Use the Team->Evaluation comment set parameter to select which set of comments you wish to use (click on the comment set link to view the list of predefined comments). Comment sets usually differ mainly in the language used, however teams might wish to create customized comment sets. Please contact the webmaster if you wish to design your own comment set.
Instructor evaluation tools
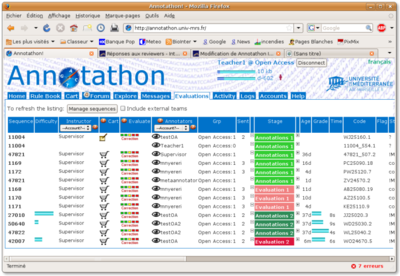
Select the Evaluations tab to display a list of the team's sequences undergoing annotation, as well as their evaluation stage (green sequences are undergoing annotation, red sequences are awaiting instructor evaluation).
Important: beware that, by default, the evaluation tab only lists the sequences assigned to your own instructor account. If your team has several instructors, you need to select Account? instead of your username in the Instructor column header popup list and press Manage sequences, to see the full list of sequences assigned to every instructor in your team.
Each line of the table corresponds to a sequence, with columns displaying:
- Sequence: Annotathon internal id.
- Difficulty: annotation difficulty as estimated by instructors.
- Instructor: username of instructor responsible for evaluating this sequence annotation.
- Cart: with particularly complex sequences, instructors can choose to add sequences to their own instructor cart, to store their own analysis results. Press the Cart icon to add this sequence to your instructor cart (then press the Edit icon to bring up the annotation form).
- Evaluate: press the Evaluation icon to mark and comment on a student annotation (see below).
- Annotators: username of students that are responsible for annotating this sequence. Press the View icon to see this student's annotations. Select a student username in the popup list in the column header and press Manage sequences to only display this student's annotation list.
- Grp: team name and subgroup number of the corresponding students.
- Sent: total number of sequence annotations submitted for evaluation, at this time, by corresponding student.
- Stage: sequence annotation stage (Annotation 1, Evaluation 1, Annotation 2, Evaluation 2, Finished). The minus and plus signs at each side of the stage can be used to force the sequence annotation stage one step back or one step forward respectively.
- Age: time elapsed since annotations were submitted by student for evaluation (in minutes, hours or days). Most urgent cases are presented first (i.e. submitted the earliest).
- Mark: marks awarded during evaluation are represented as pale blue bars (Evaluation 1) and deep blue bars (Evaluation 2).
- Time: time taken by instructor to evaluate the annotations.
- Code: annotation code, with version.
- Flag: instructor keywords to flag interesting annotation cases (see below).
- Status: coding, non coding, or !M|* (error in ORF coordinates).
- BioProc: biological process (first 15 characters).
- MolFunc: molecular function (first 15 characters).
- Gene: gene symbol.
- Conclusion: student's conclusion (first 15 characters).
- Taxonomy: student's taxonomic conclusion (first 15 characters).
- Translation: ORF protein translation (first 5 and last 5 residues).
- last update: date & time of last annotation update.
- Origin: original accession number of sequence (as provided by dataset designer).
Sequences are sorted in stage order (Annotation 1, Evaluation 1, Annotation 2, Evaluation 2, Finished), and then by decreasing submission age. Thus, annotations requiring most immediate attention (instructor evaluations pending) will appear first.
Pressing the Evaluation icon brings up the instructor marking and commenting form, for a given student annotated sequence. As soon as the form is loaded, a timer is started to help instructors manage their evaluation duties (in our experience, evaluations take between a few minutes to 30-50 minutes for complex cases).
The top two lines of this form allow the instructor to select the sequence annotation difficulty and the quantitative marks for the student annotations presented lower in the form. Student annotations are broken up into the following categories:
- ORF & molecular weight
- domains
- blast
- multiple aln
- phylogeny
- ontologies
- conclusion
Note: the annotation code at the top of the form is also a link to the standard annotation view; we have often found it useful during evaluations to open this link in another window (CTRL-click), especially if you are fortunate enough to have a dual screen display!
The fields for instructor comments are presented differently for first and second pass evaluations:
- Evaluation 1: free text comment fields, as well as tick boxes for generic predefined comments, are interleaved with the corresponding student annotations. Ticking predefined comments automatically applies a mark penalty (indicated after the tick box, e.g. -0.5) to the annotation mark, at the top of the form (mark on a 1 No results to 10 Perfect scale). These are only suggested penalties, and after reviewing all annotations, the instructor is free to select the mark he deems most appropriate.
- Evaluation 2: a single free text comment field is presented at the bottom of the form, pre-filled with a copy of the first pass instructor comments. The rationale being that, during second evaluations, the instructor essentially goes through each of the initial comments he made, and progressively deletes individual comments, if students appear to have responded adequately, in their second round of annotations. To help instructors assess changes between first and second annotations, each annotation category has a Previous version link, that unrolls the annotations, as they were initially submitted (press link again to hide). In the case of the Conclusion field, the Changes since Evaluation 1 link displays edits between first and second pass annotations in color (inserts in red, deletes in crossed out blue; press link again to hide).
If you find you are often making the same comment, when reviewing annotations, you can add a new generic predefined comment, using the New comment fields, at the end of the each evaluation form. Select the Method field carefully, as this defines where this generic comment will interleave in subsequent evaluation forms.
The Teaching flag field, at the end of the form, is provided for instructors to tag interesting annotation cases. This free text will not be displayed to students; it will only appear in the instructor Evaluation tab in the Flag column. We used this to flag up annotations we wished to discuss with class, at the beginning of practicals, or at the end of all practicals during a special debriefing lecture.
Important: after having ticked relevant generic comments, written specific comments, selected sequence intrinsic difficulty and selected the student annotation mark, press the Submit evaluations button to store the data. The annotation stage will be pushed forward, and students will be notified, by email, that the evaluation is ready. The 1-10 mark will be displayed as a proportional blue bar in their Cart tab, whilst the comments (both generic and specific) will be displayed in red at the top of their annotations (both in view and in edit modes). After submitting Evaluation 1, the annotations will be reopened to students in editing mode (Annotation 2). After submitting Evaluation 2, the annotations and associated final comments will be available to students in read only mode (Finished stage).
Account's mean intermediate marks are used to produce an instant team evaluation based ranking (bottom of Home tab). This ranking has proved useful for students to evaluate how their annotation evaluations compare with other team member's evaluations. However, it is very important that students understand that this ranking is provisional. Indeed, computation of the final grade is more subtle, taking difficulty factors into account and involving primary marks normalization, across different instructors (see below).
Computing overall grade
After all team evaluations have been completed, open the team leader Grades tab to compile the collection of marks for each student into an overall final single grade. The Grades tab has a number of computation parameters at the top of the form, press the "Grades" button to view the tentative list of grades, for each student, at the bottom of the form.
During grade computation, raw evaluation marks and difficulty factors are first normalized according to the parameters in the two first Grades->Marks & Grades->Difficulty lines of the form. These two lines are usually best left to their default values (Marks: centered on 0 / standard deviation = 3, Difficulty: centered on 4 / standard deviation=2). If your team students were asked to annotate a defined number of sequences (such as the recommended 3 sequences), then the Grades->Quality vs Quantity parameter is left at its default value of zero. If you are running a team for which no maximum limit was set to the number of sequences per student cart (such as the Open Access volunteer team), then the Grades->Quality vs Quantity parameter is used to determine the relative weight of Quality (annotation marks) versus Quantity (number of sequences annotated).
The Grades->Best sequences parameter (also modifiable from Team->Number of best evaluations to consider for grade form) is used to take only a subset of the evaluations into account during the final grade computation. For instance, if you have set the Team->Maximum number of sequences per cart to 3, and you set the Grades->Best sequences to 2, then only the two best annotations for each student will be used to compute the final grade (i.e. the worst annotation will be ignored). If set to zero (default), all available annotations will contribute to final grade.
The final Grades->Overall Grade parameter is used to renormalize the overall grades to a given average and standard deviation. Set the Grades->Overall Grade centered on and Grades->Overall Grade standard dev to appropriate values for your standard grade system. For instance, in France typical grades are on a 0 to 20 scale, so centering on 13 with a SD of 4 has often given satisfactory results.
After modifying the above parameters, press the "Grades" button, to view the tentative list of grades, for each student, at the bottom of the form. Note that a number of graphical plots and basic statistical measures are provided below the form, in order to provide some quality control estimates (final grade versus average raw marks, final grade versus subgroup etc). Comparing the separate instructor mark means/SDs, before and after normalization, should show the necessity and effectiveness of the normalization procedure.
When you are satisfied with the proposed grades, press the Submit grades button, to save the grades in the database. By default, the grades are then still confidential and only visible to the team leader. In order to let authenticated students view their final grades, as a proportional blue bar at the bottom of their Home tab, tick The grades are Public instead of Confidential, and then press the Submit grades button. Once the grades are made public, the team is listed as Closed, but students are still free to log in at any time.
The final list of student grades, at the very bottom of the Grades tab, corresponds to tab separated values, that can be copied and pasted to a standard spread sheet, for further processing.
Communication tools
Forum
The Team->Forum parameter controls whether students have access to the Annotathon internal discussion forum. When enabled, students can read posted messages, by opening the Forum tab (users are alerted to new messages, by an email and an animated icon next to the Forum tab name). It is also from this tab that students can reply to already posted messages. Forums topics roughly follow the sequence annotation fields of the Annotathon (contact the webmaster to have more topics added):
- General Annotathon issues, using the interface, bugs etc.
- ORF finding
- Homolog hunting: BLAST
- Multiple sequence alignements
- Phylogenetic analysis
- Ontologies (molecular function & biological process)
- Conserved domains
- Conclusion
However, to post new messages, students must always use the form provided at the top of their sequence annotations in view mode (Cart tab->eye icon, as described in the Rule Book). This ensures that, when posting questions, students have their annotations handy and that posted forum questions contain a direct link to the sequence annotations. It is indeed essential for those wishing to reply to a message (usually instructors, but also students) that they have access to the sequence annotations concerned. In order to allow posting of messages of general interest, that are not linked to a specific sequence (such as analysis hints, of interest to all), instructors can post new messages directly from their Forum tab page (special instructor form at the bottom of the Forum page).
Different sets of forum topics can be selected using the Team->Forum set parameter (click on the forum set name, to see the list of forum topics); forum topic sets usually differ according to language used. By default, the Forum tab only displays messages issued by same team members. By ticking the Forum->Include messages from teams other than own team, messages from all teams, using the same forum set will be displayed.
Chat
You can enable or disable the Annotathon internal chat, using the Team->Chat parameter. When enabled, authenticated Annotathon users have an extra Chat tab which allows instant messaging, between logged in users of the same team. The chat can prove useful to promote communication between students outside classes; however the chat can prove distracting during 'real' practical sessions, supervised by instructors, and it is recommended you disable the chat service just before class and re-enable afterwards. Note that for instructors, the Chat is by default set to PRIVATE, whereby instructor chat exchanges are private to the team's instructors (students excluded). The instructor chat can be switched to PUBLIC mode, by ticking the bow in the top right hand corner of the Annotathon page (chat messages are then visible to students).
Instructor announcements
Instructors have access to the Messages tab, which is used to make either team wide announcements or send private messages to specific team members. Once posted, the message is displayed prominently, at the top of each Annotathon page of the recipients, until they tick the Read box to the left of the message. The message is then archived, for future reference, in the recipient's messages archive, at the bottom of their Forum tab. Note that students can not send such direct messages, they only use the public Forum so that questions and answers are shared by all participants.
Customizing the set of student annotation fields
In the Team->Annotation fields section, tick the fields your students will be required to fill-in, in the sequence annotation editing form. The currently available annotations are:
- ORF fields (ORF finder results, start & stop locations, strand)
- Coding/non-coding status
- Molecular weight estimation
- Conserved protein domains
- BLAST report
- BLAST taxonomy report
- Multiple sequence alignment
- Phylogenetic tree
- GO Biological Process
- GO Molecular Function
- Gene symbol
- Taxonomic classification
- Conclusion field
- Note pad
If you would like to see other fields added to this list, please contact the webmaster.
Results fields template
To help student better structure their various results sections, we have found it beneficial to apply the following standard template to all Annotation fields:
PROTOCOL ----------------------------------------------------------------------------------------------------- RESULTS ANALYSIS ----------------------------------------------------------------------------------------------------- RAW RESULTS -----------------------------------------------------------------------------------------------------
You can change this template in the Team->Results fields template section.
Conclusion field maximum length
To encourage students to keep their conclusion to the point (and begin practicing the art of filling out grant application forms), you can enter an integer value in the Team->Conclusion field maximum length box. Students will then be presented with a warning when the number of characters in their conclusion is over the Team->Conclusion field maximum length value. We have found around 3000 letters is a reasonable default for the conclusion alone.
Gene Ontology controlled vocabularies
Use the Team->Ontology set parameter to select what limited list of Gene Ontology terms students of your team will be allowed to choose from, in the Biological Process and Molecular Function fields of the annotation forms. Click on the Team->Ontology set link to unfold the corresponding list of GO terms.
Our first source of DNA fragments was an ongoing yeast genome project, therefore the first Team->Ontology set corresponds to a yeast GO slim subset. For metagenomic sequences, we have now added a more appropriate second set, corresponding to the GO prokaryote subset. If you wish to create a new set, please contact the webmaster.
Wiki export
Team leaders have access to the Export tab, that allows export of student annotations to a public external Wiki. Annotations that are not exported remain private to Annotathon registered and authenticated users only; web search engines such as Google will therefore never index Annotathon data that is not exported. Annotations are exported at the team leader's discretion, usually after final grades have been computed, by default to the Metagenes Wiki (based on the Wikipedia engine). The Metagenes annotations are then public, both for reading and editing, by any interested party.
The Export tab lists student annotations in order of descending marks. Each annotation has a tick box which determines which ones are set for potential export. By default, annotation with a mark of at least 7/10 are ticked. This minimum value can be changed in the Export->Auto-selection of annotations with over box and by pressing Select. Annotations can, of course, also be selected by hand.
After the relevant annotations have been ticked, press the Wiki button to start exportation to Metagenes. The export process can be very lengthy, please do not interrupt the process once started. Exported annotations can not be retracted from inside the Annotathon (they can be deleted from inside Metagenes). Annotations can be re-exported, overwriting existing Metagenes pages (each annotation is exported to a page named after the Annotathon annotation code).
Bioinformatics Analyses
Overview
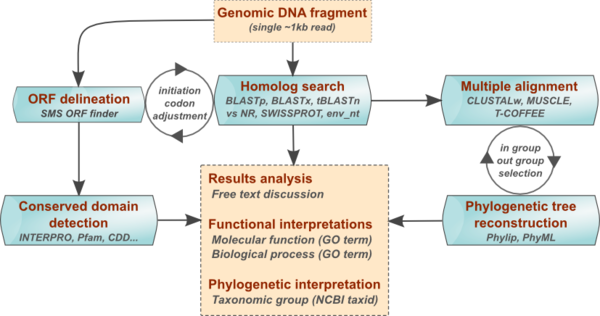
The annotation process involves the following main steps:
- ORF identification
- Determination of protein molecular weight
- Research of conserved protein domains
- Research of sequence homologies
- Multiple alignments of homologs
- Building of a phylogenetic tree
- Identification of taxonomic group
- Sequence annotation with Gene Ontology
- Proposition of a gene symbol
- Writing of synthetic conclusions
Depending on the DNA sample, several iterations of these analyses steps may be needed to derive consistent annotations, for example refining ORF delineation, in the light of BLAST results, or yet improving multiple alignments to obtain better phylogenetic trees (cf Figure Annotation work flow opposite).
You can use the Team->Annotation fields check boxes to select which of the above analyses will be applicable to your team. For instance, if you don't want your students to be presented with the Gene symbol field, just untick the appropriate Gene symbol box.
Rule Book & FAQ (Frequently Asked Questions)
Before they can start Bioinformatics analyses, annotators must have been introduced to the Rule Book and must have loaded at least one DNA sample into their cart. The Rule Book provides the practical background necessary for each of these steps, along with links to websites providing additional information. A Help tab lists a series of links to webpages providing further information on relevant topics (e.g. Tree of Life), tool suites such as Sequence Manipulation Suite (SMS) or bioinformatics resource centers (NCBI, EBI, etc.). The Help tab can be overlayed, at any time, on any Annotathon page, by clicking on the Help tab, with links usually opening in other windows, in order to preserve current student work in progress forms.
As soon as students start with bioinformatics analyses per se, they should constantly refer to the Frequently Asked Questions (FAQ) which is linked in the Help tab at the top of each page. The FAQ, which should be kept open in a separate browser window or tab at all times, provides numerous hints, warnings, tutorials and screen shots for all the required bioinformatics analyses. The FAQ notably contains essential information about phylogenetic tree inference (e.g. study group selection strategies, and common pitfalls of tree interpretation).
Bioinformatics tools
All analyses can be performed with freely available software, either using local software or distant web servers. A list of the recommended tools is provided in Table Bioinformatics tools used by students. Fore most analysis steps, we propose several alternative tools, which can typically be used, with different options or parameters (cf. fourth column of Table Bioinformatics tools used by students).
| Analyses | Tools | Websites | Variants, options & parameters | Results |
|---|---|---|---|---|
| ORF delineation | SMS ORF Finder | http://annotathon.org/sms2/ | With/out start codons | Putative ORF coordinates, translation, initiation and stop codons |
| NCBI ORF Finder | http://www.ncbi.nlm.nih.gov/gorf/gorf.html | 2x3 frames | ||
| EBI ORF Finder | http://www.ebi.ac.uk/Tools/emboss/transeq/index.html | With/out start codons | ||
| Molecular weight | SMS suite | http://annotathon.org/sms2/protein_mw.html | Default | Molecular weight |
| Conserved domains | Interpro | http://www.ebi.ac.uk/Tools/InterProScan/ | Default | Putative domains and associated molecular functions |
| Pfam | http://pfam.jouy.inra.fr/ | Default | ||
| ProSite | http://www.expasy.org/prosite/ | Default | ||
| Homolog search | BLASTp | http://blast.ncbi.nlm.nih.gov/Blast.cgi | Databases: SWISSPROT | Summary of Blast output file Selection of putative homologous sequences (Fasta format) |
| tBALSTn | http://www.ebi.ac.uk/Tools/blastall/index.html | Non redundant protein sequences (NR) | ||
| BLASTx | http://www.expasy.org/prosite/ | Environmental samples (env_nt) | ||
| Multiple alignments | CLUSTALw | http://www.ebi.ac.uk/Tools/clustalw2/index.html | Ouput format (CulstalW) | ClustalW formated multiple alignment (Multiple Alignment Annotathon field) |
| MUSCLE | http://www.ebi.ac.uk/Tools/muscle/index.html | |||
| T-COFFEE | http://www.ch.embnet.org/software/TCoffee.html | |||
| Phylogenetic trees | Phylip | http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py | Neighbor-joining (NJ) | Phylogenetic tree with properly annotated branches (protein names, species acronyms) |
| PhyML | http://www.phylogeny.fr/ | Maximun likelyhood | ||
| Taxonomic classification | NCBI taxid | http://www.ncbi.nlm.nih.gov/Taxonomy/ | NCBI numerical identifier or scientific name (to fill in specific boxes) | |
| Functional annotation | GO terms | http://www.geneontology.org/ | Molecular functions & Biological processes | GO terms to fill in specific boxes |
As they proceed with their analyses, apprentice annotators fill out specific fields in the dedicated Annotathon sequence editing form, which is automatically controlled for coherency with predefined types. This form has two types of fields:
- (i) raw results (e.g. as is outputs from ORF finders, BLAST, multiple alignments & phylogenetic trees);
- (ii) interpretations and conclusions (i.e. molecular functions, biological processes, gene symbol, taxonomy & conclusions).
Thus, the Annotathon editing form constitutes both a numeric "laboratory note book" (type 1 fields) and an "annotation report" (type 2 fields). Some brief contextual help is provided for each annotation field of the editing form, by clicking the question mark icons. Finally, a "Notepad" field (at the bottom of the sequence editing form) allows student to store temporary or intermediate data, or conclusions awaiting further processing. This space is private to students and should not be consulted during evaluation.
According to our experience, the most delicate steps are the selection of an appropriate set of homologous sequences, the construction of an informative multiple alignment, and the construction of a coherent phylogenetic tree. Each of these steps may thus require specific briefings, at appropriate times, by the teacher. We provide below some troubleshooting information on specific steps.
Delineation of Open Reading Frame
- Rule number 1: if there are more than one decent sized ORFs, then students should study the longest available ORF (and totally ignore the smaller ones)
- Rule number 2: the ORF end position should not include the STOP codon (i.e. the end position is the last codon coding for an amino acid)
- Depending on the type of ORF finding web site used, students may have some trouble finding the longest ORF in their sequence. Make sure all frames are explored.
- Filling the “ORF start” “ORF end” fields is a problem when ORFs are on the reverse strand. In this case, students have to count positions from the 3’ end and they may have to try adding +1 or -1 to the result to find the real start/stop positions. Using the SMS Orf finder tool is recommended as the ORF start and stop positions reported by this tool are in the correct coordinates. The only exception is that if the ORF is 3' complete (contains the STOP codon), then students need to subtract 3 from the end position.
- Be aware that many DNA samples do not contain a full coding sequence. When specified ORFs lack an ATG or Stop codon, the system displays a message such as “Info: the sequence does not start with ATG/end with a Stop codon”. There is nothing you can do about it, except reminding students about the limitations of shotgun sequencing! Note that you must continue the annotation session, for this sequence, even though it may not contain a full gene.
Protein Functional Roles
- For retrieving protein homologs, a good procedure is to start with Blastp against Swissprot. If no significant (or too few) homologs are found, then widen the search using the “nr” database.
- Sequence selection for multiple alignment and phylogenetic tree is a tricky part, leading to many discussions during the session. When several protein functions are present among Blast hits, then we suggest sampling these functions reasonably (ie no more than 3-4 functions in a tree), and using the phylogenetic analysis to assign the most likely function to the query protein.
- In the best case scenario, the phylogenetic tree will show distinct ortholog groups, with the query protein located among one group. This enables unambiguous functional assignement. Unfortunately, a number of events may spoil this picture. First, many protein names in databases are just inferred, by homology, and, as a result, may be described as members of family X, while indeed they belong to family Y. As a result, families X and Y will be mixed up in the final tree. The same kind of problem arises, when different names are used for a single protein family, which is very frequent. Finally, some trees are poorly resolved, due, for instance, to the presence of highly divergent sequences in the set. In such cases, functional assignment must be achieved directly from Blast results, assuming the query protein’s function is that of the highest scoring Blast hit. However, students must be informed of the dangers of such an interpretation.
- On the basis of the functional annotations associated with the homologs and protein domains found, the apprentice annotator has to select the most appropriate GO terms to describe the ORF under evaluation, focusing on the biochemical activity of the protein (Molecular Function) and on its role in the cell (Biological Process). These terms should be inferred from the closest homologs or most significant domains found.
- Only once convincing homology has been found, with a family of well characterized proteins of known function, can a Gene symbol be proposed for the ORF. The corresponding fields should be kept empty, if the closest homologs have no associated Gene symbol, or if the homology relationships are ambiguous.
Taxonomic Classifications
- On the basis of the phylogenetic tree produced, the most likely taxonomic group can be deduced. This group can be filled in on the annotation form, by specifying the NCBI numerical identifier found in GENBANK records, in the feature table, or by querying the NCBI taxonomy database. Alternatively, the exact scientific name for this group can be pasted in the 'Scientific name' box.
- Once the annotations have been saved, the field left blank should be correctly, automatically, populated. The "NCBI numerical identifier" box has precedence over the "Scientific Name" box. Thus, to change the taxonomic classification of the current ORF, the numeric code must be deleted, before entering a new "Scientific Name".
- The precision of the taxonomic classification should reflect the quality of the phylogenic tree obtained.
Links, References & Contacts
- Annotathon portal: http://annotathon.org/
- Instructor manual: http://annotathon.org/Metagenes/index.php/Annotathon_Instructor_Manual
- Student user guide: http://annotathon.org/?actionjs=rules
- About the Annotathon: http://annotathon.org/Metagenes/index.php/Annotathon
- Announcements & Discussions : http://groups.google.com/group/annotathon
- Source code: https://launchpad.net/annotathon
- Contact: Pascal Hingamp