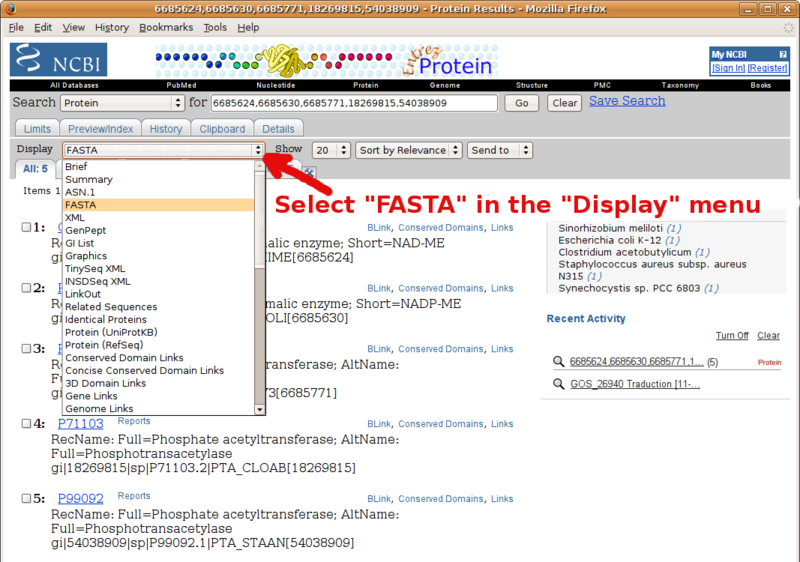
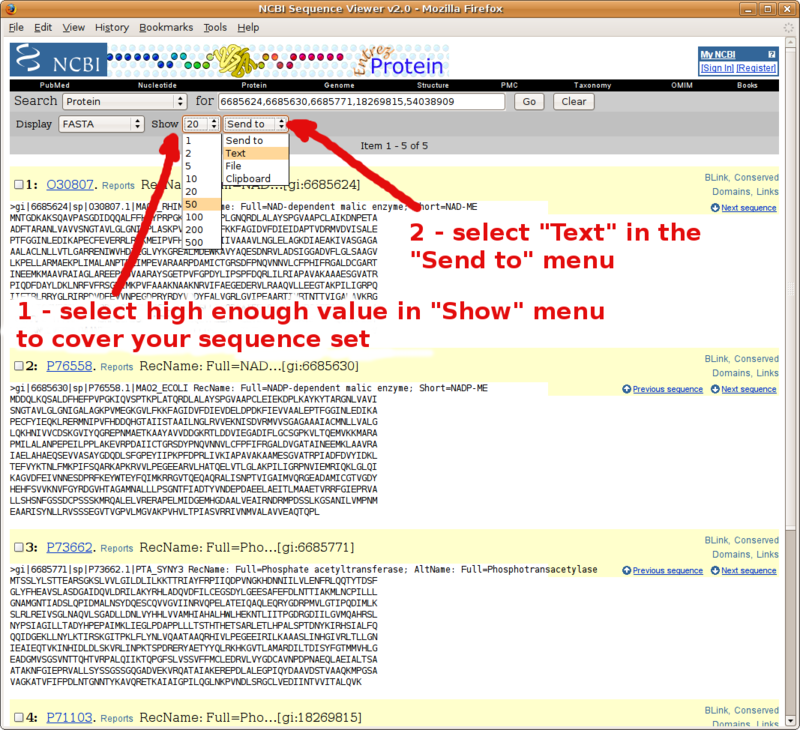
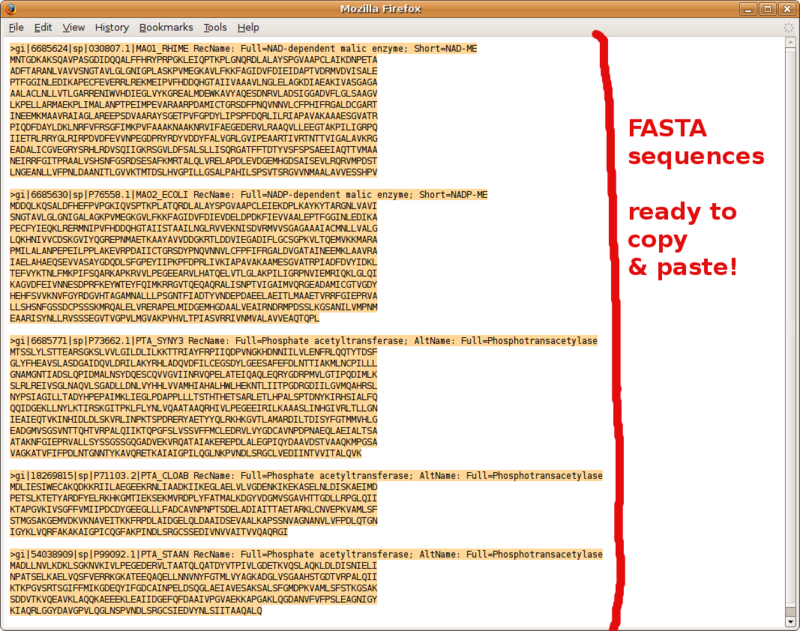
Frequently Asked Questions
This FAQ presents in depth explanations of the bioinformatics analyzes necessary for the Annotathon. For a explanations on how to create user accounts and general sequence management issues, please refer to the user manual, also called the Rule book. Finally, please note this is a Wiki, so everyone is invited to contribute to this documentation!
Contents
[hide]- 1 Translation, ORFs and coding/non-coding status
- 2 INTERPRO: Identifying conserved protein domains
- 3 BLAST: Finding sequence homologs
- 4 Using BLAST to compile a list of FASTA formatted sequence homologs
- 5 A microbial view of the Tree Of Life
- 6 Designing sequence ingroups and outgroups for phylogenetic tree inference
- 7 Phylogeny.fr: Infering phylogenetic trees
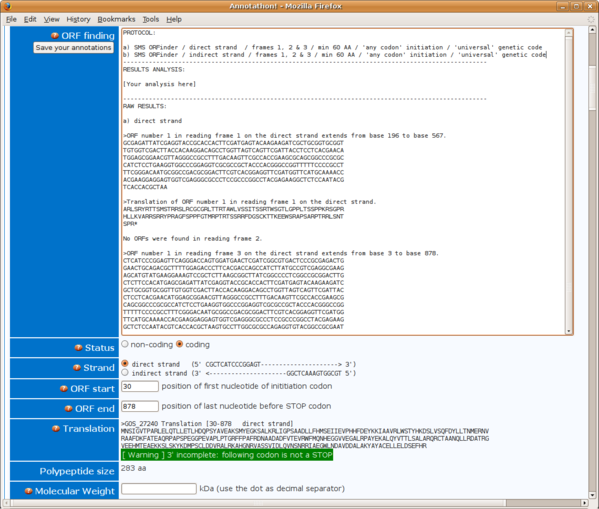
Translation, ORFs and coding/non-coding status
- If the genomic DNA does not contain any Open Reading Frames (ORFs, here defined as a stretch of at least 60 codons without a single STOP codon), then immediately conclude NON-CODING (tick non-coding under STATUS). Other than a very brief word of conclusion, no other analyses or annotations are required (non coding DNA annotation is difficult with very short random metagenomic DNA reads).
- If the genomic DNA does contain ORFs over 60aa in length, proceed with the rest of the analysis with the longest available ORF. Two outcomes are possible:
- analysis of the longest ORF shows homologs and/or conserved protein domains => select coding STATUS and proceed with the rest of the analyzes (multiple alignements, phylogeny etc.)
- analysis of the longest ORF shows no homologs (even in the ENV_NR environmental sequence database) and no conserved protein domains => discuss if the DNA is coding or not and select the appropriate STATUS. If the ORF is very long (say over 200aa), then it is likely that this ORF does indeed code for a protein: it is then called an ORFan - an ORF with no known homologs! If the ORFan is only just above the 60aa length threshold, you might want to classify it as non-coding. Also beware of low complexity DNA (e.g. repeated stretches of the same bases), as this is often found to yield long false positive ORFs (in which case the translations usually also bear highly prominent AA repeats). In any case, discuss your choice and always carry out a BLASTx before concluding that the DNA is non-coding. Only proceed with the analysis of a lesser sized ORF, if it is largely overlapping with a longer ORFan and shows BLAST homologs or conserved protein domains. This is not so common but has been seen a few times in the GOS data (the real ORF, with clear homology to known proteins, is contained in a larger false positive ORF with no matches, usually antisense).
As far as what inititation codon parameter to select in the ORF finding software, start with the greedy approach: that which produces the longest possible ORFs (i.e. use "any codon" for ORF start in SMS/Orfinder). If later on your multiple alignments seem to suggest that all your homologs start further downstream, then revisit the ORF start position by locating its most likely start codon (the one closest in position to the homolog's starts).
In terms of what genetic code to use for generating the ORF, select either the "universal/standard code", or the one most likely used by the hosts of your DNA fragments ("bacterial" for marine samples passed through .8 micron filters).
If you use SMS/ORFinder, remember to carry out the analysis in all 6 frames! Frames 1, 2 & 3 on direct then indirectstrand.
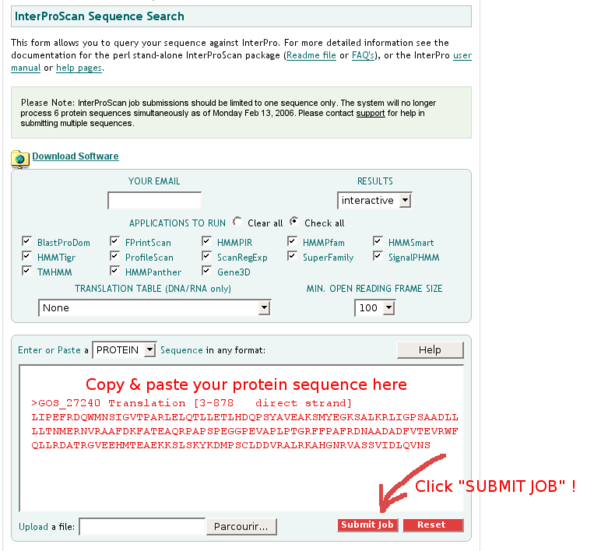
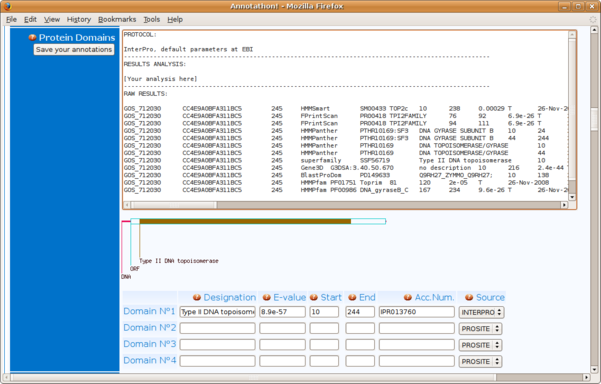
INTERPRO: Identifying conserved protein domains
Identifying conserved protein domains in a protein is a powerful method to predict its putative function.
Paste your protein sequence in the sequence field of the InterproScan tool: click "Submit Job" to scan your protein against the large InterPro federation of protein domain databases.
You might have to wait a few minutes for the results to be returned. In the resulting list of predicted InterPro domains, take note of the following points:
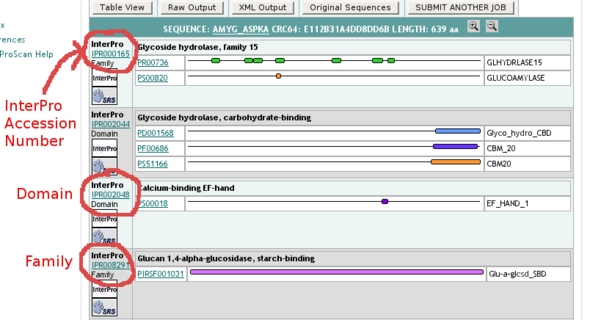
- an InterPro record (e.g. IPR000165) corresponds to a number of identified conserved domains from the underlying databases (here domain PR00736 from PRINTS & PS00820 from PROSITE); please indicate in the corresponding Annotathon field the InterPro Accession Number (here IPR000165)
- all the InterPro domains identified in your sequence are not necessarily independent: some domains might be contained in others, or can have child/parent relationships. Click on "Table View" at the top of the results page to obtain a more detailed output (including domain start & stop positions, as well as the all important E-values associated with the predictions)
- ignore any InterPro domains that are tagged as "Unintegrated", unless you have absolutely nothing else to feed your "functional role" line of investigation
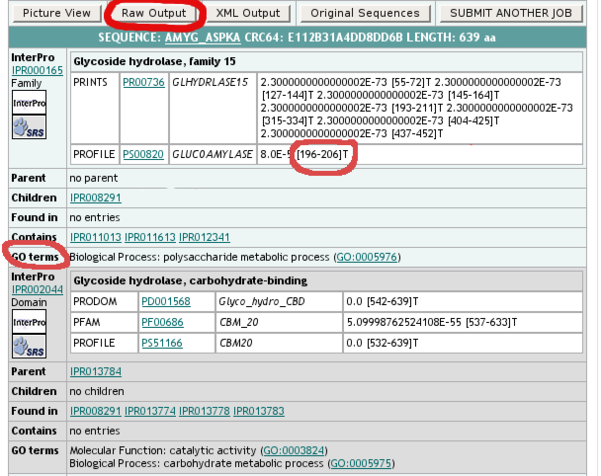
- click on the "Raw output" button to see the full results in the "text only" format, suitable for copying & pasting into the "Domains" raw results section of the Annotathon (copy the results in extenso, not just the domains your consider of interest)
You can see which other domains are linked to the domains identified in sequence in the "Children"/"contains"/"found in" sections of the InterPro scan results. Rule: only define a specific domain in the Annotathon for the largest encompassing domain, i.e. the domain which contains the other ones.
In this example, the first domain (IPR000165) has domains IPR008291 as children (4th in the results list); in this case only indicate IPR000165, and not IPR008291.
It is also in the Table View that you will find the exact coordinates of the predicted domains; for domain IPR000165, report the extremities of the PROSITE domain, and not those of all the small PRINTS sub-fragments!
Note that in some InterPro records, you will find precious functional hints for the conserved domains. You can use these functional indications to help you select appropriate Gene Ontology terms for your protein (Molecular Function and/or Biological Process). Sometimes, specific GO terms are in fact directly associated with InterPro domains, which might prove very useful if you think these GO annotations can be transfered to your particular protein.
BLAST: Finding sequence homologs
Search for known protein sequences that look similar to your ORF (potential homologs) by running BLAST, preferably using the NCBI online BLAST (since it presents the all important Taxonomy Report), or at other institutions offering online BLASTs (e.g. the EBI, or GigaBlaster@IGS).
The most usual BLASTs for the Annotathon are:
- BLASTp versus SWISSPROT: to find homologs that are well annotated (e.g. molecular functions etc.)
- BLASTp versus NR: find all possible homologs (e.g. to carry out a phylogenetic analysis)
- BLASTx versus NR: translates your genomic fragment directly into the 6 possible frames and then runs 6 BLASTp's (if you are unsure of the ORF location, or if you suspect sequencing errors producing frameshifts)
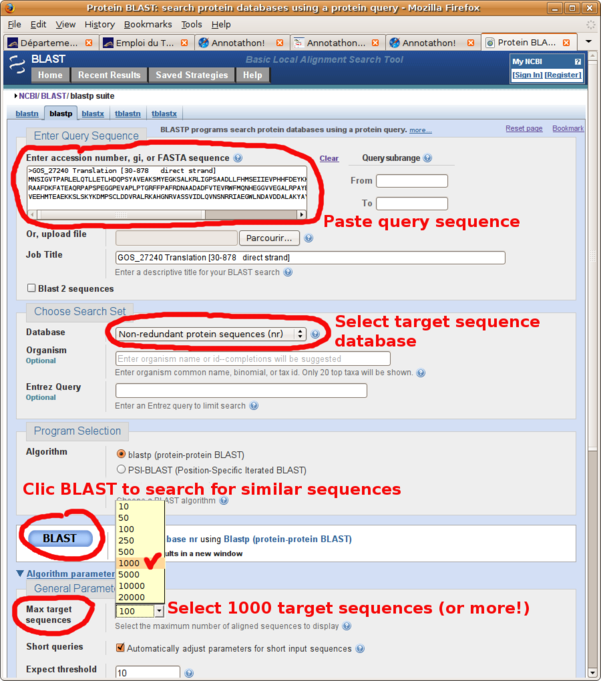
Start by filling out the BLAST online form(Fig. B1):
- copy/paste your query sequence (ORF protein sequence for BLASTp, full genomic DNA sequence for BLASTx)
- select the databank you wish to search: usually SWISSPROT or NR (NR is a compilation of all protein databanks, and therefore contains all known protein sequences)
- select a higher number of Max target sequences than the default 100 (say 1000, sometimes more) in order to get the full spectrum of homologs. If you don't select a high enough number of target sequences, then your list of similar sequences might end up truncated (you will know this is the case when the bottom of your resulting list of BLAST hits doesn't reach the default E-value threshold of 10).
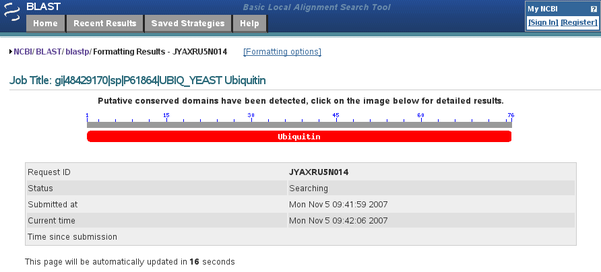
After having submitted the search, wait until the 'BLAST Status ... Searching' page (Fig. B2) is finally replaced by the results page. Note, however, that this 'BLAST Status ... Searching' intermediate page can sometimes present a colored diagram which corresponds to a conserved domain search result (in this case against the CDD domain database). This can prove very useful (see above), but has nothing to do with the BLAST results per se!
For a BLAST result which you wish to report in the Annotathon, please always include in the Annotathon BLAST section (Fig. B6):
- a header/protocol line which non-ambiguously describes what search was carried out (ex: BLASTp versus SWISSPROT, NCBI default parameters other than "500 max target sequences")
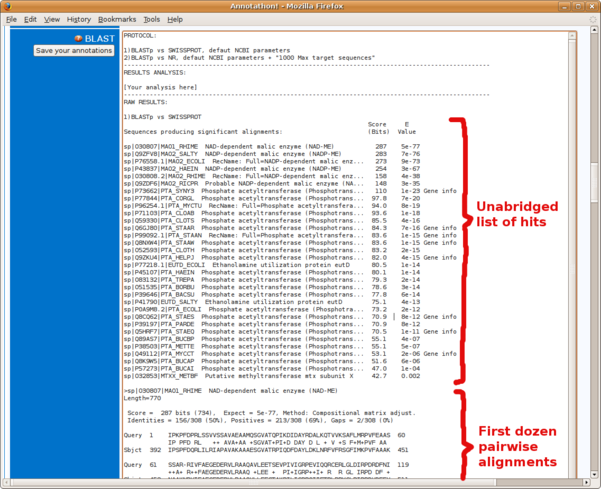
- the full, unabridged, list of hits and E-values (Fig. B4)
- the first dozen pairwise alignments only (Fig. B5)
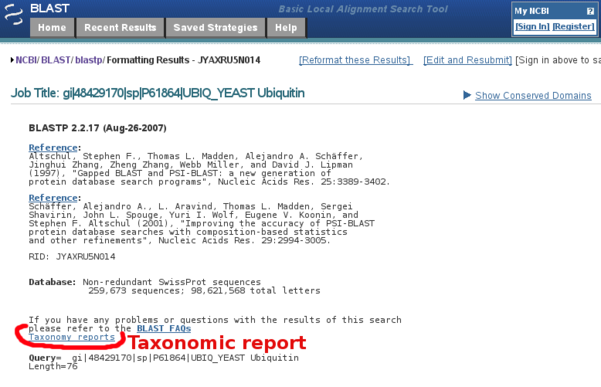
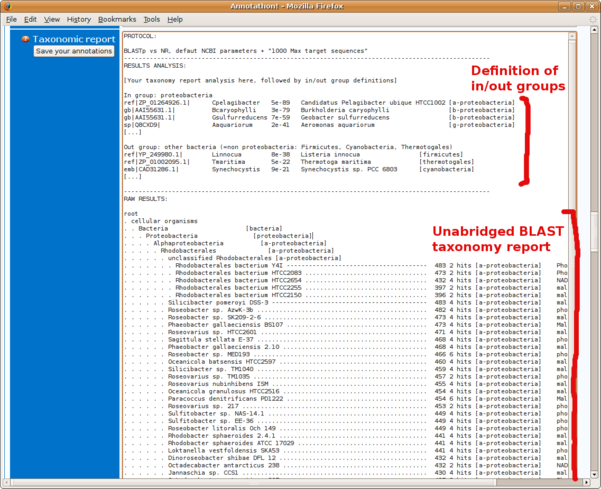
- the full, unabridged, taxonomic report (the first section, entitled Lineage Report Fig B7) copied into the Annotathon "Taxonomic Report" section (Fig B8)
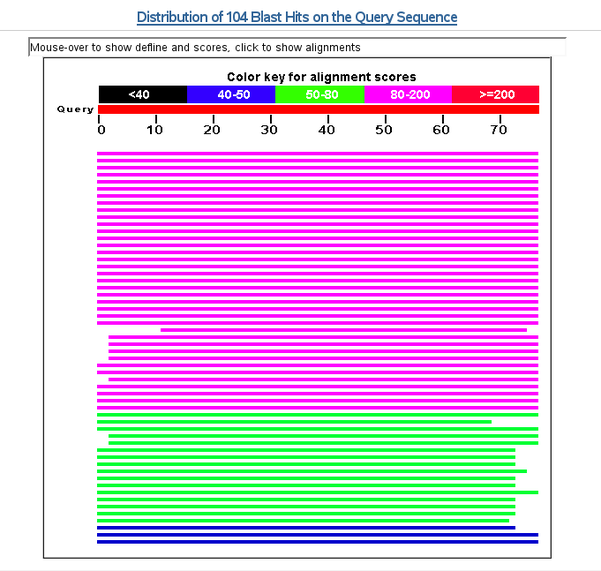
NCBI graphical overview of pairwise alignments found
List of BLAST hits and E-values
List of detailed BLAST pairwise alignments
Annotathon: BLAST results section
NCBI BLAST "Taxonomic Report" (Lineage report)
If you wish to report more than one BLAST results in the Annotathon
(e.g. one vs SWISSPROT & one versus NR), copy them one after the
other in the Annotathon field with a line of dashes as a
separator(-----------------------------).
Using BLAST to compile a list of FASTA formatted sequence homologs
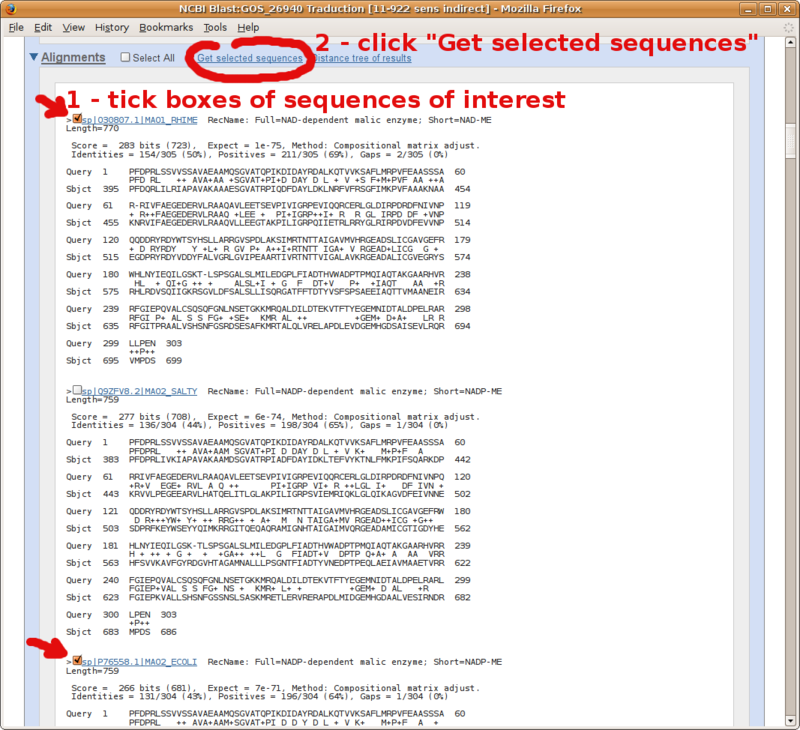
Take advantage of your fresh BLAST main results page to compile a set a FASTA formatted sequences (e.g. your in group and out group sequences to carry over to phylogenetic analysis).
Go to the pairwise alignments section of your NCBI BLAST report, and follow instructions in the following screenshots.
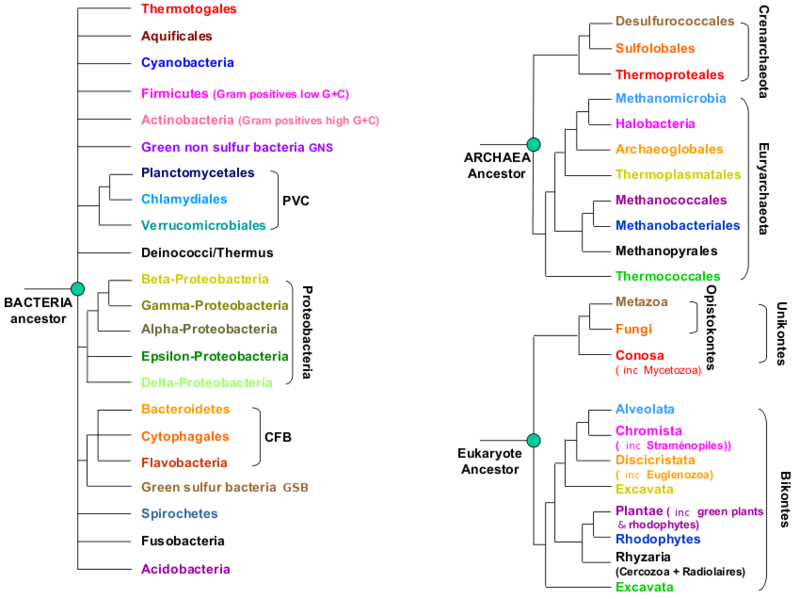
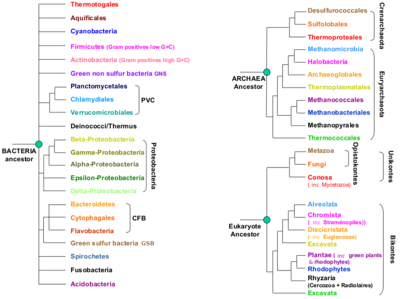
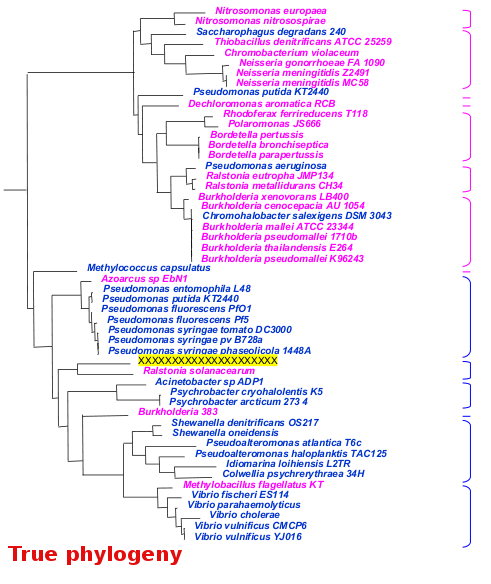
A microbial view of the Tree Of Life
It is essential that metagenomic sequence annotators keep this simplified Tree of Life within reach at all time! Understanding the branching patterns is quintessential to correctly define in- and out-groups for infering phylogenetic trees. You could print out the image below, or make it your Desktop background image...
Designing sequence ingroups and outgroups for phylogenetic tree inference
Strategy for defining ingroups and outgroups
- Define the ingroup so that it represents a true taxonomic lineage
- It must be a monophyletic group!
- Choose a wide enough range of sequences so that all ingroup lineages are represented
- Define the outgroup as the set of all other lineages of the same taxonomic level as the ingroup
- Choose a wide enough range of sequences so that all outgroup lineages are represented
- If you have no available sequence homologs for outgroup species, then you will have no outgroup (the tree will be unrooted)
Above all, remember that each and every sequence you wish to include in your phylogenetic tree should be a clear homolog of the others: each sequence should have a credible BLAST E-value when aligned to your query, and each sequence must fit snuggly in the multiple sequence alignment! Any sequence that looks like it doesn't belong to the same family, or is too partial (truncated) compared to other members of the family, should be removed from the in or out groups!
Valid examples of ingroups (always refer to the Tree Of Life):
- Cyanobacteria
- Thermotogales
- delta-Proteobacteria
- Planctomycetales + Chlamidiales + Verrucomicrobiales (PVC)
- Proteobacteria
- beta-Proteobacteria +gamma-Proteobacteria
- Bacteria
- Archaea
- Cellular organisms (Archaea + Bacteria + Eukaryotes)
Invalid ingroups:
- Cyanobacteria + Firmicutes
- alpha-Proteobacteria + beta-Proteobacteria
Example n°1:
- Ingroup = Firmicutes
- Outgroup = other lineages of same taxonomic level (i.e. all other bacterial phyla: Thermotogales, Aquificales, Cyanobacteria, Proteobacteria, PVC...)
- Your groups should contain representatives of each of these phyla
Example n°2:
- Ingroup = alpha-Proteobacteria
- Outgroup = other lineages of same taxonomic level (i.e. all other Proteobacteria: beta, delta, epsilon, & gamma)
- Your groups should contain representatives of each of these classes
Example n°3:
- Ingroup = gamma-Proteobacteria
- Outgroup = other lineage of same taxonomic level (i.e. beta-Proteobacteria)
- Your groups should contain representatives of each of these two classes
Example n°4:
- Ingroup = Bacteria
- Outgroup = other lineages of same taxonomic level (i.e. Archae & Eukaryotes)
- Your groups should contain representatives of each of these domains
Example n°5:
- Ingroup = Bacteria
- If there are no archeal or eukaryotic homologs, you will not use an outgroup (the resulting tree will be unrooted!)
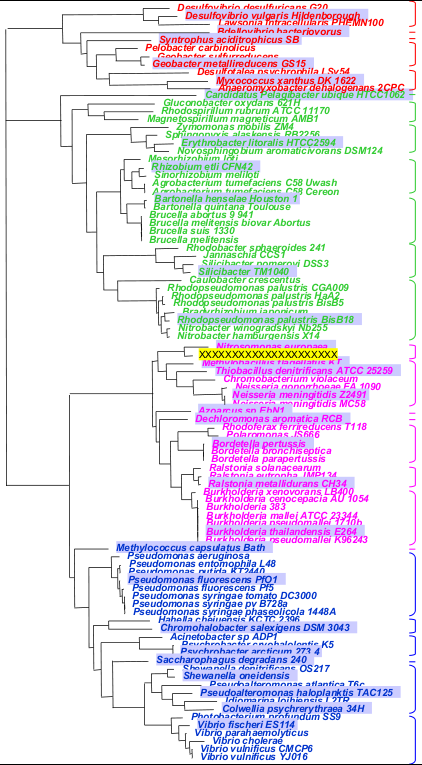
Important: it is essential that you select the sequences to build the in and out groups in such a way that these group's full diversities are well represented (i.e. that you have sequence representatives of each of the subgroups that make up the in and out groups). Use the above simplified Tree Of Life, the NCBI Taxonomy browser and the BLAST Lineage Report to identify existing subgroups.
Example: In the tree opposite, the in group is made up of the pink and blue branches, the unknown query sequence is highlighted in yellow. The out group is made of the green and red branches.
- for the in group, pick 15-30 sequences in each one of the subgroups enclosed by a pink or blue bracket (e.g. those on a grey background)
- for the out group, pick 5-10 sequences in each one of the subgroups enclosed by a red or green bracket (e.g. those on a grey background)
- in the example across, the resulting phylogeny would successfully suggest that the query sequence belongs to the pink group, probably even to the same subgroup as Nitrosomonas europaea
- under no circumstance should you just pick the set of first 15 best BLAST scoring hits for you in (or out) group! This will usually result in just representing a single subgroup...
List of complete microbial genomes at NCBI
| Bacteria | |
| Gamma-Proteobacteria | 145 |
| Firmicutes | 129 |
| Alpha-Proteobacteria | 79 |
| Beta-Proteobacteria | 48 |
| Actinobacteria | 48 |
| Cyanobacteria | 30 |
| Epsilon-Proteobacteria | 19 |
| Delta-Proteobacteria | 18 |
| Bacteroidetes/Chlorobi | 17 |
| PVC | 13 |
| Spirochaetes | 9 |
| Chloroflexi | 8 |
| Thermotogales | 6 |
| Thermus/Deinococci | 4 |
| Acidobacteria | 2 |
| Aquificales | 1 |
| Other | 2 |
| Archaea | |
| Euryarchaeota | 33 |
| Crenarchaeota | 15 |
| Nanoarchaeota | 1 |
To the right is a table of complete microbial genomes are available at the NCBI (2007):
Of course, many more partial genome sequences from other bacteria or archae are present in GENBANK or SWISSPROT. However, if a study conducted with a gamma-proteobacterial in group reveals only a handful gamma-proteobacteria, then uttermost care is required during interpretation. Indeed, since over 145 complete gamma-bacterial genomes are available in GENBANK, this might indicate horizontal gene transfers, or massive gene loss in this group.
Common pitfalls & difficulties in building trees
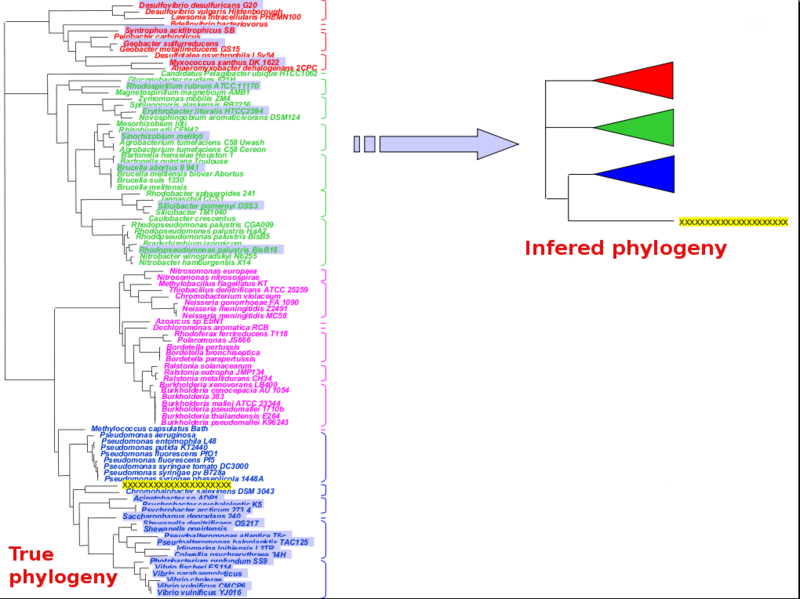
Pitfall 1: in group sequences do not fully represent group diversity
In the figure across, the in group is made of the pink + blue groups, the query sequence is highlighted in yellow, and the out group is made of the red + green groups.
An incorrect selection of in group sequences is indicated by the light grey backgrounds. The resulting inferred phylogeny (right panel) will show the query sequence emerging exactly between the in group and the out group. This usually indicates :
- an incorrect selection of sequences to represent the in group (and/or out group).
- an incorrect definition of the in group (and/or out group)
- Solution 1: select a more rational set of sequences that better represents the in group
- Solution 2: redefine the in and out groups
- If no amount of wider ranging in group sequence selection manages to integrate the query sequence, then this might be a true biological signal rather than an artefact (see below). This does arise occasionally when dealing with metagenomes since these sequences can come from uncultured bugs belonging to potentially never seen before taxonomic subgroups (i.e. discovery?).
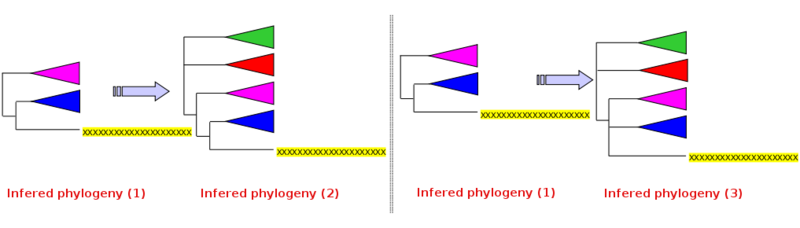
Difficulty 1: query sequence never integrates the in group
In the figure below, the in group is blue, the outgroup pink, and the query sequence yellow (infered phylogeny n°1). Regardless of the efforts to properly represent the in group diversity, the query sequence always emerges between the in and out groups. Left in this state, no conclusion is possible from the infered phylogeny n°1.
- Solution : broaden the out group (add further green and
red out groups) and rerun tree inference. Two outcomes are possible
using the broadened outgroup:
- the query sequence is specifically linked to the in group (without integrating the latter, inferred phylogeny n°2): it is legitimate to conclude that the query sequence is a close relative of the in group, even if one can not conclusively state that it is part of it. The query sequence represents either an unknown subgroup of the in group, or it represents an unknown novel group, close relative of the in group
- the query sequence is not specifically linked to the in group in particular (inferred phylogeny n°3): it is legitimate to conclude that the query sequence represents an unknown altogether novel group, not specifically related to the in group
Difficulty 2: anomalous classifications of in and out group sequences (HGT's)
In the figure opposite, sequence classification according to the inferred phylogeny presents occasional contradictions with the accepted reference phylogeny (Tree of Life). Some in group sequences are mixed in the out group branch, and/or outgroup sequences are mixed within the in group branch. Less dramatic anomalies occur when in and outgroup sequences are well separated, but mixes occur between lineages within either the in group or the outgroup.
- Explanation : the sequence is likely to be subject to horizontal gene transfers (HGT's, some genes are more frequently observed in HGT's, such as antibiotic resistance genes and various transporters). In the figure opposite, we can only conclude that the sequence is a close relative of Ralstonia solanacearum, without it being possible to assign the query to either pink or blue groups.
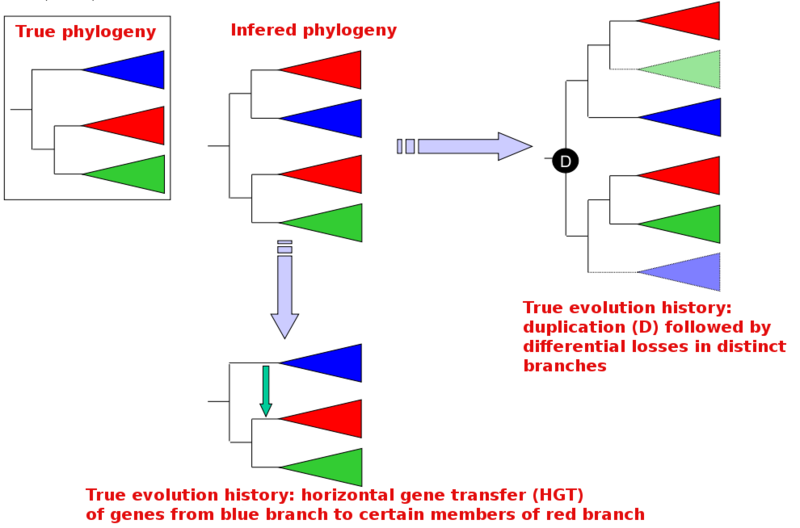
Difficulty 3: anomalous classifications of in and out group sequences (duplications)
In the figure below, the conventional species phylogeny is shown on the left (True phylogeny). The phylogeny inferred from a set of homologous sequences is shown in the center (Inferred phylogeny), and shows an additional red branch linked specifically to the blue branch. This unexpected inferred phylogeny can be explained by either:
- Gene duplication followed by differential losses in various lineages (right panel)
- Horizontal gene transfer from the blue branch to some members of the red group (bottom panel)
Resolving past duplication events is notoriously difficult; it usually involves restricting the analysis to species for which a complete genome sequence is available, allowing the inference of trees containing all paralogs and orthologs involved. However, differential gene loss which often follows gene duplications can make inferred trees rather cryptic...
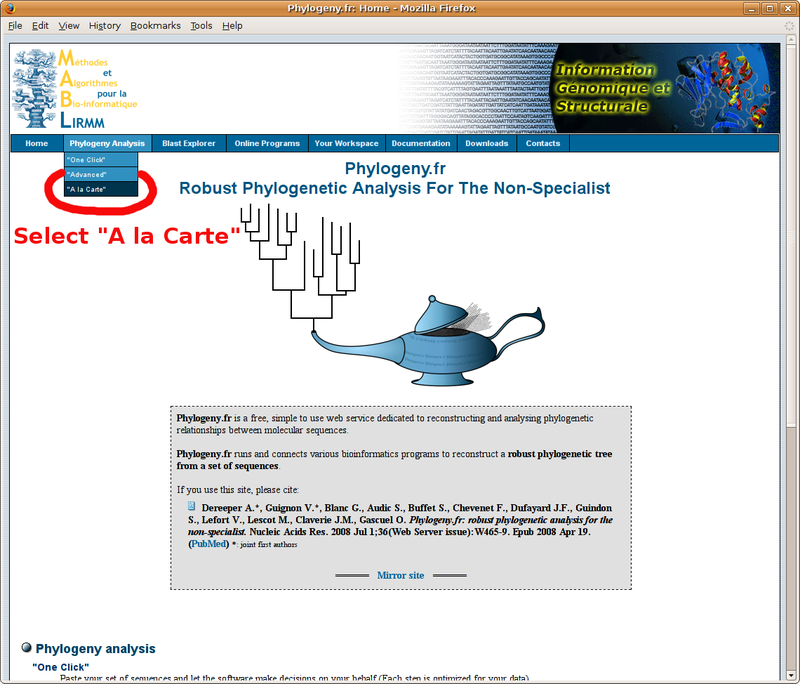
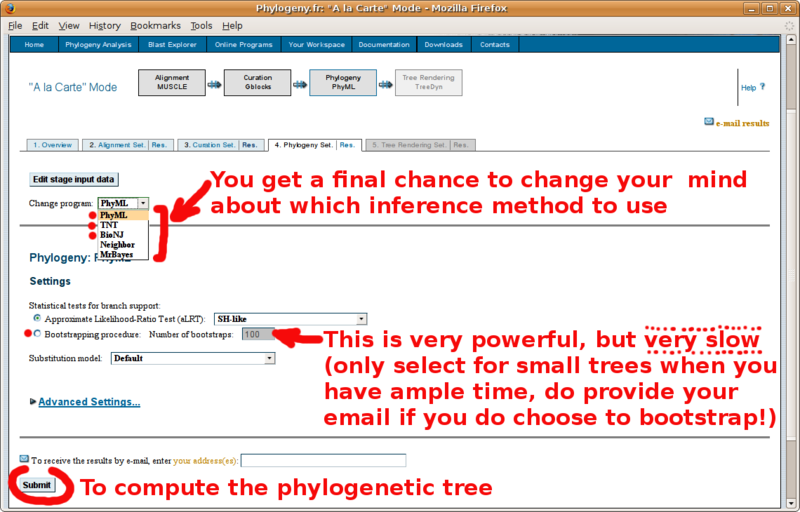
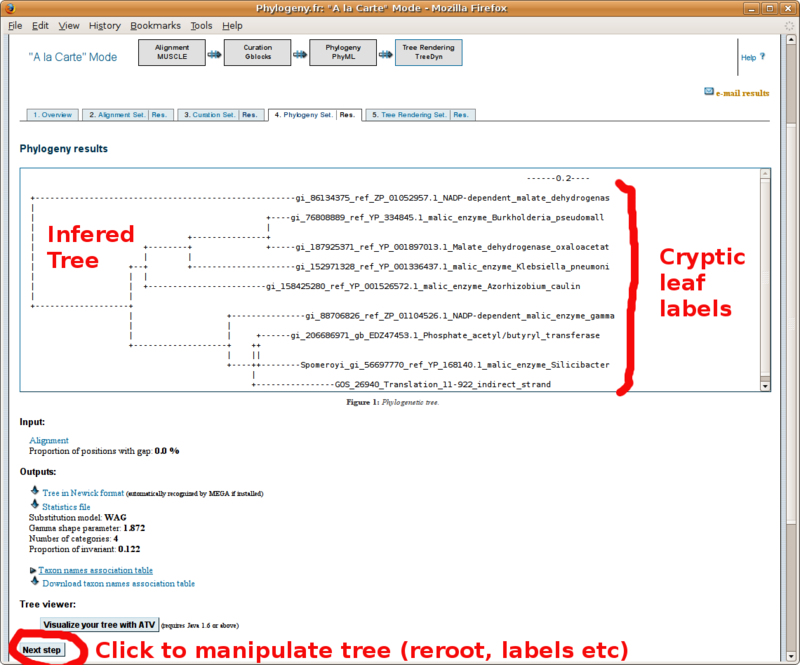
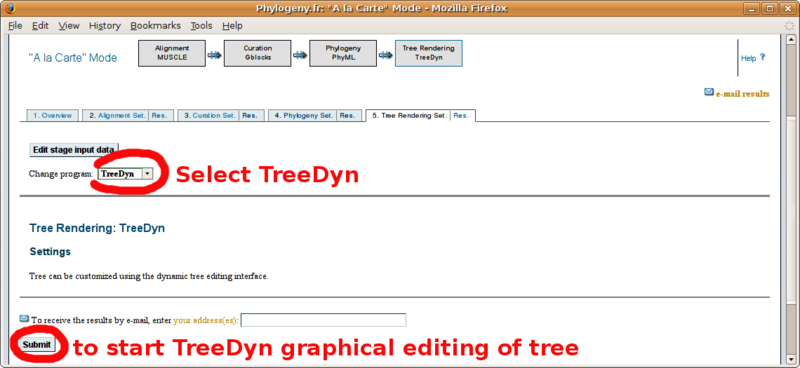
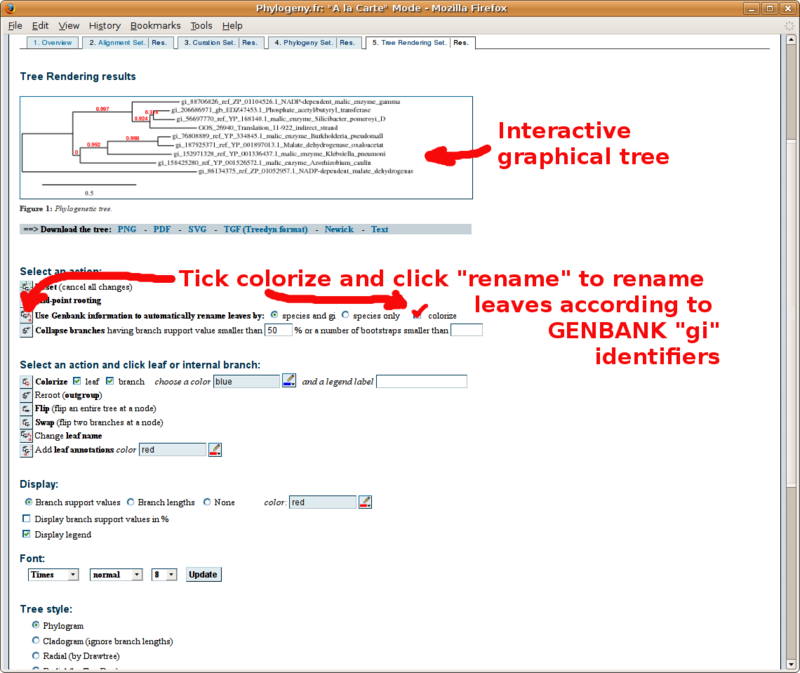
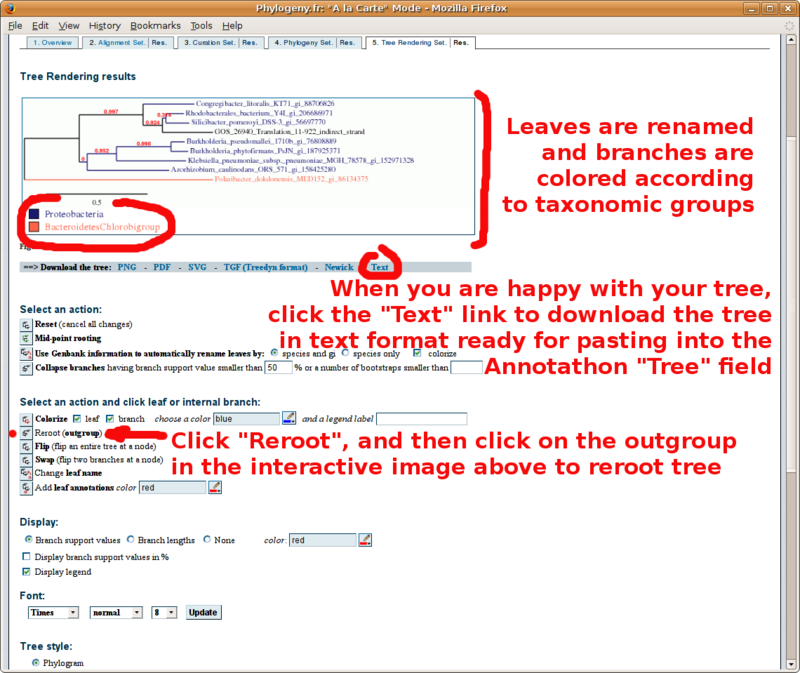
Phylogeny.fr: Infering phylogenetic trees
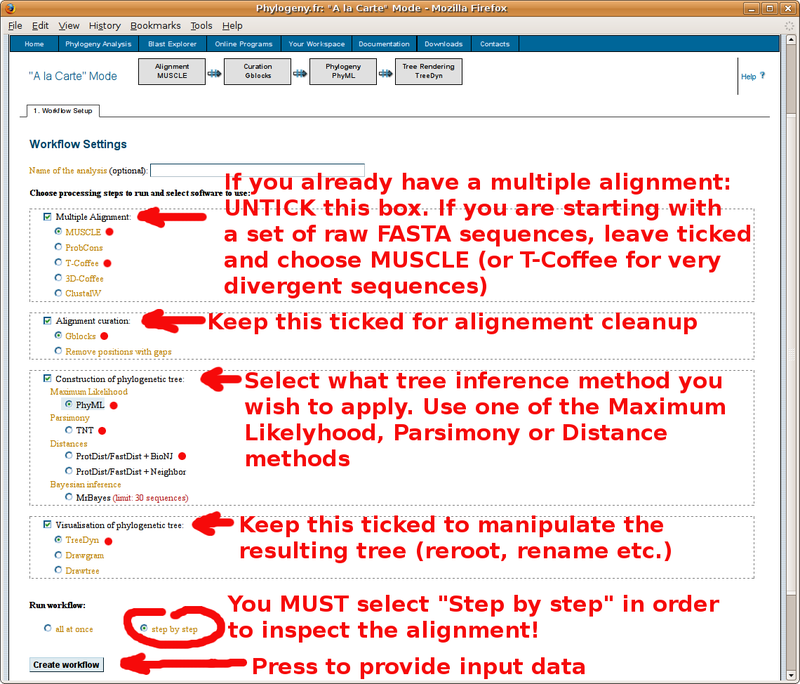
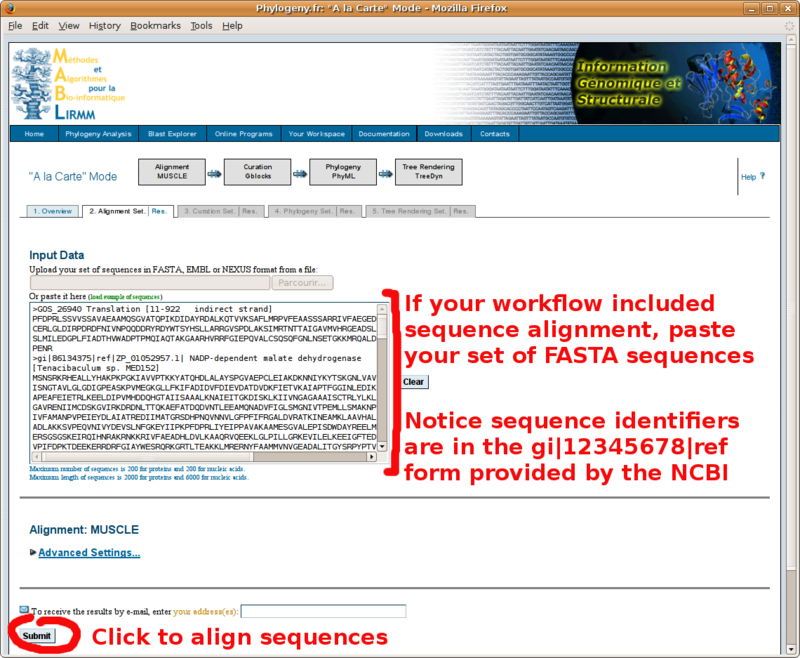
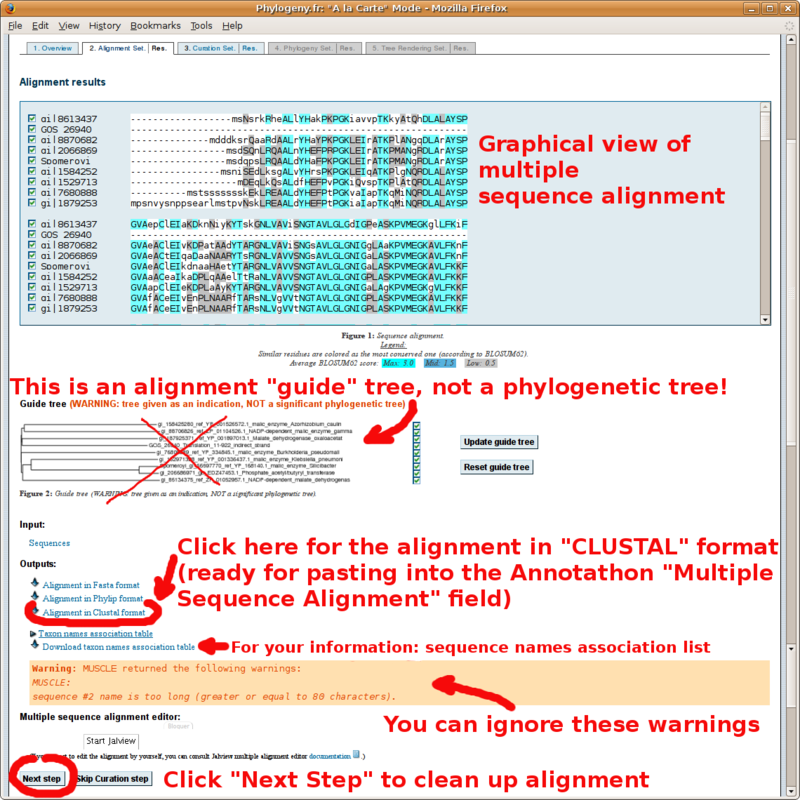
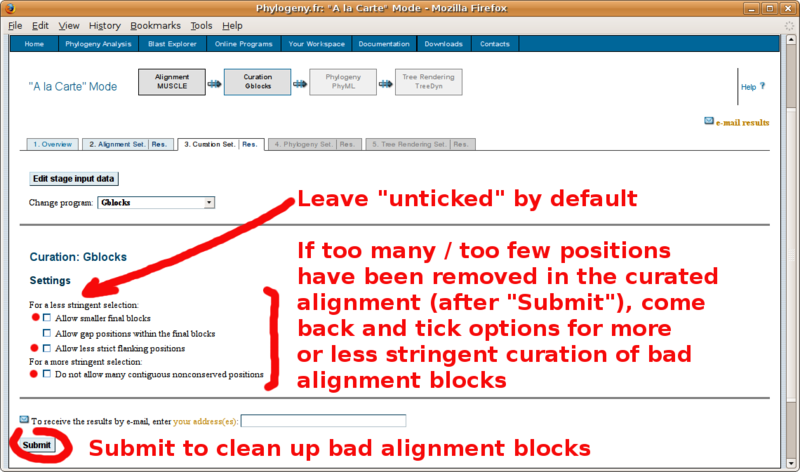
With your in-group and out-group set of FASTA formatted sequences, point your browser to the http://www.phylogeny.fr/ online site for multiple sequence alignment and phylogenetic tree construction. You will find below a screenshot tutorial of the full procedure:
The "MUSCLE" format alignment obtained by clicking the "Alignment in CLUSTAL format" link (paste in Annotathon multiple alignment field):
):
MUSCLE (3.7) multiple sequence alignment
gi|8613437 ------------------MSNSRKRHEALLYHAKPKPGKIAVVPTKKYATQHDLALAYSP
GOS_26940 ------------------------------------------------------------
gi|8870682 --------------MDDDKSRQAARDAALRYHAYPKPGKLEIRATKPLANGQDLARAYSP
gi|2066869 -----------------MSDSQNLRQAALNYHEFPRPGKLEIRATKPMANGRDLARAYSP
Spomeroyi -----------------MSDQPSLRQAALDYHAFPKPGKLEIRATKPMANGRDLARAYSP
gi|1584252 ----------------MSNISEDLKSGALVYHRSPKPGKLEIQATKPLGNQRDLALAYSP
gi|1529713 -------------------MDEQLKQSALDFHEFPVPGKIQVSPTKPLATQRDLALAYSP
gi|7680888 ----------MSTSSSSSSSKEKLREAALDYHEFPTPGKVAIAPTKQMINQRDLALAYSP
gi|1879253 MPSNVYSNPPSEARLMSTPVNSKLREAALDYHEFPTPGKIAIAPTKQMINQRDLALAYSP
gi|8613437 GVAEPCLEIAKDKNNIYKYTSKGNLVAVISNGTAVLGLGDIGPEASKPVMEGKGLLFKIF
GOS_26940 ------------------------------------------------------------
gi|8870682 GVAEACLEIVKDPATAADYTARGNLVAVISNGSAVLGLGNIGGLAAKPVMEGKAVLFKNF
gi|2066869 GVAEACTEIQADAANAARYTSRGNLVAVVSNGSAVLGLGNIGALASKPVMEGKAVLFKNF
Spomeroyi GVAEACLEIKDNAAHAETYTARGNLVAVVSNGTAVLGLGNIGALASKPVMEGKAVLFKKF
gi|1584252 GVAAACEAIKADPLQAAELTTRANLVAVVSNGTAVLGLGNIGPLASKPVMEGKAVLFKKF
gi|1529713 GVAAPCLEIEKDPLAAYKYTARGNLVAVVSNGTAVLGLGNIGALAGKPVMEGKGVLFKKF
gi|7680888 GVAFACEEIVENPLNAARFTARSNLVGVVTNGTAVLGLGNIGPLASKPVMEGKAVLFKKF
gi|1879253 GVAFACEEIVENPLNAARFTARSNLVGVVTNGTAVLGLGNIGPLASKPVMEGKAVLFKKF
gi|8613437 AMKLAAVHALADLAKKSVPEQVNIVYDEVSLNFGKEYIIPKPFDPRLIYEIPPAVAKAAM
GOS_26940 -----------------------------------------PFDPRLSSVVSSAVAEAAM
gi|8870682 AMQLACIDGIAALSRATTSAEAAEAYRGEQLVFGVDYLIPKPFDPRLMGVVASAVASAAM
gi|2066869 EMQIACVDGIAELARATTSAEAAAAYKGEQLNFGADYLIPKPFDPRLVAVVSSAVAKAAM
Spomeroyi AMQIACVEGIAELARITTSAEAAAAYQGEQLTFGADYLIPKPFDPRLVGVVSSAVARAAM
gi|1584252 EMKMAAVEAIAALARETPSDVVARAYGGETRAFGADSIIPSPFDPRLILRIAPAVAKAAM
gi|1529713 EMKLAAVHAIAELAHAEQSEVVASAYGDQDLSFGPEYIIPKPFDPRLIVKIAPAVAKAAM
gi|7680888 EMEIAAVNAIAELAQQEQSDIVATAYGIQDLSFGPEYLIPKPFDPRLIVKIAPAVAQAAM
gi|1879253 EMEIAAVNAIAELARQEQSDIVATAYGIQDLSFGPEYLIPKPFDPRLIVKVAPAVAKAAM
****** :..*** ***
gi|8613437 ESGVALEPISDWDAYREELMERSGSGSKEIRQIHNRAK---RNKKRIVFAEADHLDVLKA
GOS_26940 QSGVATQPIKDIDAYRDALKQTVVKSAFLMRPVFEAAS---SSARRIVFAEGEDERVLRA
gi|8870682 ETGVATRPVEDLVAYRERLDASVFRSSMIMRPVFAAAA---LSQRRIVFAEGEDERVLRT
gi|2066869 ESGVATRPIEDITAYKQKLNQTVFKSALLMRPVFEAAR---AAARRIVFAEGEDERVLRA
Spomeroyi ESGVARRPITDLEAYRQKLNQSVFKSALLMRPVFEAAA---KAARRLVFAEGEDERVLRA
gi|1584252 DTGVATRPIADFDAYNEKLDEFVFRSGFIMRPLFQRAK---QDKKRVIYAEGEDERVLRA
gi|1529713 DSGVATRPIADFDAYIEKLSEFVYKTNLFMKPIFSQAR---KEPKRVVLAEGEETRVLHA
gi|7680888 DGGVATRPIEDMEAYKVHLQQFVYHSGTTMKPVFQIARGAPAEKKRVVFAEGEEERVLRA
gi|1879253 DSGVAERPIEDMEAYEQHLQQFVYHSGTTMKPIFQLARGVEPEKKRIVFAEGEEERVLRA
: *** *: * ** * :. :. * .*:: **.: **.:
gi|8613437 AQRVQEEKLGLPILLGRKEVILELKEEIGFT----EDVPIFDPKTDEEKERRDRFGIAYW
GOS_26940 AQAVLEETSEVPIVIGRPEVIQQRCERLGLDIRPDRDFNIVNPQQD---DRYRDYWTSYH
gi|8870682 AQVIVEEMTDRPILIGRPEIIARRCEKAGLTIKPGEDFEVVNPEDD---SRHRRYWEAYL
gi|2066869 AQAILEETTETPILIGRPEVIERRCEKLGLDVRPGRDFQLVNPEND---PRYYDYWNSYH
Spomeroyi AQAILEETTETPILIGRPEVIEARCEKMGLSVRPGQDFQIVNPEND---PRYYDYWTSYH
gi|1584252 AQAVIEEGIAHPILVARPSVLEARLQRFGLSIRPGKDFEVINPEDD---PRYRDFVRSYI
gi|1529713 TQELVSLGLAKPILVGRPSVIEMRIQKLGLQIKAGVDFEIVNNESD---PRFKEYWSEYY
gi|7680888 VQIVVDEKLAKPILIGRPAVIEHRIQRYGLRLTPGVDFTIVNTEHD---ERYRDFWQTYF
gi|1879253 MQIIVDEKLAKPILIGRPAVIEQRIARYGLRLIAGQDYTVVNTDHD---ERYRDFWQEYH
* : . **::.* :: *: * :.: . * * : *
gi|8613437 ESRQRKGRTLTEAKKLMRERN-YFAAMMVNVGEADALITGYSRPYPTVIRPILESIQKDS
GOS_26940 SLLARRGVSPDLAKSIMRTNTTAIGAVMVHRGEADSLICGAVGEFRWHLNYIEQILGSK-
gi|8870682 QLMSRRGVTPDLAKVIMRTNTTAIAAIMVYCGDADSMVCGSFGQYLWHLNYVRQILAYD-
gi|2066869 KVMQRRGVTPDLAKAIMRTNTTAIGAIMVHRGEADSLLCGTFGEYRWHLNYVQQVLGGG-
Spomeroyi QLMERRGVTPDIAKAIMRTNTTAIGAIMVHRGEADSLICGTFGEYRWHLNYVEQVLGSK-
gi|1584252 EIAGRRGVTPDAARTLVRTSSTVISALAVKKGEADAMLCGIEGRFSRHLRHVRDIIGLAP
gi|1529713 QLMKRRGITQEQAQRAVISNTTVIGAIMVHRGEADAMICGTIGEYHDHYRVVQPLFGYRD
gi|7680888 KMMARKGISEQLARVEMRRRTTLIGSMLVKKGEADGMICGTISTTHRHLHFIDQVIGKRA
gi|1879253 KMMSRKGISAQMAKLEMRRRTTLIGAMLVEKGEADGMICGTVSTTHRHLHFIDQVIGKKE
. *.* : *. : . :.:: * *:**.:: * . : :
gi|8613437 GISKVAACNLMLTKQGPMFLADTTINLNPTAKDLVKISQMTSNLVKMFGMKPNVAMLSFS
GOS_26940 TLSPSGALSLMILEDGPLFIADTHVWADPTPMQIAQTAKGAARHVRRFGIEPQVALCSQS
gi|8870682 GAHPRGALSLMITEDEPLFIADTHVHPEPTPEQIADTVMAAANHVRRFGMKPNIALCSHS
gi|2066869 TYSPHGALSMMILEDGPLFIADTHVHVEPTPEQIAETVIGAARHVRRFGLAPKIALCSQS
Spomeroyi DLRPHGALSLMILEDGPLFIADTHVRSRPSPEELAEITLGAARHVRRFGIEPQIALCSQS
gi|1584252 GVRELAALSLLITPKGNLFLCDTQVQTEPNAADLAEMTILAAAHVRRFGIEPKVALLSHS
gi|1529713 GVSTAGAMNALLLPSGNTFIADTYVNHDPSPEELAEITLMAAESVRRFGIEPRVALLSHS
gi|7680888 GCSVYGAMNALVLPGRQIFLVDTHVNVDPTPAQLAEITIMAAEEVRRFGIEPKVALLSHS
gi|1879253 GAKVYAAMNALVLPNRQIFLVDTHVNVDPTPEQLAEITIMAAEEVRRFGIEPKIALLSHS
.* . :: *: ** :. *.. ::.. :: *. **: *.:*: * *
gi|8613437 NFGSTKNESSQKIREAVSYIHRNFPNAVVDGEIQADFALNPEMLAKEFPFSKLNGKKVNV
GOS_26940 QFGNLNSETGKKMRQALDILDTEKVTFTYEGEMNIDTALDPELRARLLPENR--------
gi|8870682 QFGNLDIDSGRRVRQAMALLEAREPDFAYEGEMHIDSALDPDLRARIFPNSRLQG-PANV
gi|2066869 QFGNISCDTGSRLRAAIEILDDKRRDFVYEGEMNIDTALDPELRERIFPNSRLEG-AANV
Spomeroyi QFGNQAEGSGQRLRQAIEILDSRPRDFVYEGEMNLDSALDPELRQRIFPNSRLYG-AANV
gi|1584252 NFGSNDTVCARRVRAALDILKDRAPELEVDGEMQAELALLPDARERILPHSRLQG-VANV
gi|1529713 NFGSADCPSASKMRKTLELVKARAPELMIDGEMHGDAALVESIRNDRMPDSPLKG-AANI
gi|7680888 NFGTSNAPSAQKMRDTLAILQERAPDLHVDGEMHGDVALDAALRKEILPESTLEG-EANL
gi|1879253 NFGTSNAPTAQKMRDTLAILRERAPDLQVDGEMHGDIALDANLRREVMPDSTLEG-DANL
:**. . .:* :: : :**:: : ** :* .
gi|8613437 LIFPNLESANITYKLLKEMQG-AESIGPVILGLSKAVHIVQLGASVDEMVNMAALACVDA
GOS_26940 ------------------------------------------------------------
gi|8870682 LVFAYGDAASGVRNILKMRGG-ALEVGPILMGMGNRAHIVTPSITARGLLNISALAGTDV
gi|2066869 LIFAHADAASGVRNILKMRAG-GLEVGPILMGMGNRAHIVSPSITARGLLNMAAIAGTPV
Spomeroyi LIFAHADAASGVRNVLKMKAN-GIEVGPILMGMGNRAHIVTPSITARGLLNMAAIAGTPV
gi|1584252 LVMPDLDAADIAYNMIKVLGD-ALPVGPILMGTAKPAHILGPTVTARGIVNMTAVAVVEA
gi|1529713 LVMPNMEAARISYNLLRVSSSEGVTVGPVLMGVAKPVHILTPIASVRRIVNMVALAVVEA
gi|7680888 LVLPNIDAANIAYNLLKTAAGNNIAIGPILLGAAQPVHVLTESATVRRIVNMTALLVADV
gi|1879253 LVLPNIDAANISYNLLKTAAGNNIAIGPMLLGAAKPVHVLTASATVRRIVNMTALLVADV
gi|8613437 QQREKK
GOS_26940 ------
gi|8870682 THYS--
gi|2066869 AHYG--
Spomeroyi AHYG--
gi|1584252 QSEA--
gi|1529713 QTEPL-
gi|7680888 NAVR--
gi|1879253 IAAR--
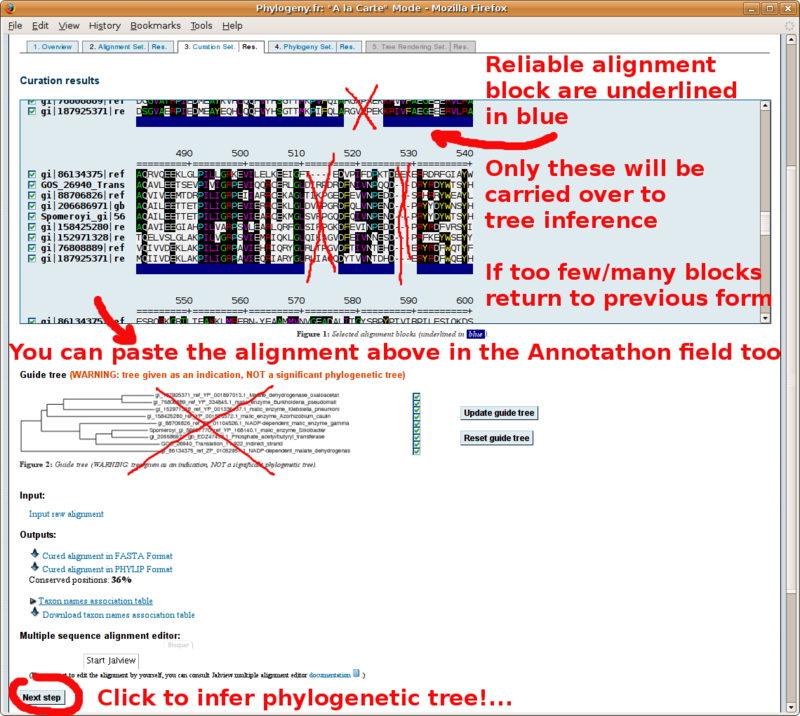
The GBLOCKS curated multiple sequence alignment (paste in Annotathon multiple alignment field):
Gblocks 0.91b Results
Processed file: input.fasta
Number of sequences: 9
Alignment assumed to be: Protein
New number of positions: 288 (selected positions are underlined in blue)
10 20 30 40 50 60
=========+=========+=========+=========+=========+=========+
gi|86134375|ref ------------------MSNSRKRHEALLYHAKPKPGKIAVVPTKKYATQHDLALAYSP
GOS_26940_Trans ------------------------------------------------------------
gi|88706826|ref --------------MDDDKSRQAARDAALRYHAYPKPGKLEIRATKPLANGQDLARAYSP
gi|206686971|gb -----------------MSDSQNLRQAALNYHEFPRPGKLEIRATKPMANGRDLARAYSP
Spomeroyi_gi|56 -----------------MSDQPSLRQAALDYHAFPKPGKLEIRATKPMANGRDLARAYSP
gi|158425280|re ----------------MSNISEDLKSGALVYHRSPKPGKLEIQATKPLGNQRDLALAYSP
gi|152971328|re -------------------MDEQLKQSALDFHEFPVPGKIQVSPTKPLATQRDLALAYSP
gi|76808889|ref ----------MSTSSSSSSSKEKLREAALDYHEFPTPGKVAIAPTKQMINQRDLALAYSP
gi|187925371|re MPSNVYSNPPSEARLMSTPVNSKLREAALDYHEFPTPGKIAIAPTKQMINQRDLALAYSP
70 80 90 100 110 120
=========+=========+=========+=========+=========+=========+
gi|86134375|ref GVAEPCLEIAKDKNNIYKYTSKGNLVAVISNGTAVLGLGDIGPEASKPVMEGKGLLFKIF
GOS_26940_Trans ------------------------------------------------------------
gi|88706826|ref GVAEACLEIVKDPATAADYTARGNLVAVISNGSAVLGLGNIGGLAAKPVMEGKAVLFKNF
gi|206686971|gb GVAEACTEIQADAANAARYTSRGNLVAVVSNGSAVLGLGNIGALASKPVMEGKAVLFKNF
Spomeroyi_gi|56 GVAEACLEIKDNAAHAETYTARGNLVAVVSNGTAVLGLGNIGALASKPVMEGKAVLFKKF
gi|158425280|re GVAAACEAIKADPLQAAELTTRANLVAVVSNGTAVLGLGNIGPLASKPVMEGKAVLFKKF
gi|152971328|re GVAAPCLEIEKDPLAAYKYTARGNLVAVVSNGTAVLGLGNIGALAGKPVMEGKGVLFKKF
gi|76808889|ref GVAFACEEIVENPLNAARFTARSNLVGVVTNGTAVLGLGNIGPLASKPVMEGKAVLFKKF
gi|187925371|re GVAFACEEIVENPLNAARFTARSNLVGVVTNGTAVLGLGNIGPLASKPVMEGKAVLFKKF
370 380 390 400 410 420
=========+=========+=========+=========+=========+=========+
gi|86134375|ref AMKLAAVHALADLAKKSVPEQVNIVYDEVSLNFGKEYIIPKPFDPRLIYEIPPAVAKAAM
GOS_26940_Trans -----------------------------------------PFDPRLSSVVSSAVAEAAM
gi|88706826|ref AMQLACIDGIAALSRATTSAEAAEAYRGEQLVFGVDYLIPKPFDPRLMGVVASAVASAAM
gi|206686971|gb EMQIACVDGIAELARATTSAEAAAAYKGEQLNFGADYLIPKPFDPRLVAVVSSAVAKAAM
Spomeroyi_gi|56 AMQIACVEGIAELARITTSAEAAAAYQGEQLTFGADYLIPKPFDPRLVGVVSSAVARAAM
gi|158425280|re EMKMAAVEAIAALARETPSDVVARAYGGETRAFGADSIIPSPFDPRLILRIAPAVAKAAM
gi|152971328|re EMKLAAVHAIAELAHAEQSEVVASAYGDQDLSFGPEYIIPKPFDPRLIVKIAPAVAKAAM
gi|76808889|ref EMEIAAVNAIAELAQQEQSDIVATAYGIQDLSFGPEYLIPKPFDPRLIVKIAPAVAQAAM
gi|187925371|re EMEIAAVNAIAELARQEQSDIVATAYGIQDLSFGPEYLIPKPFDPRLIVKVAPAVAKAAM
###################
430 440 450 460 470 480
=========+=========+=========+=========+=========+=========+
gi|86134375|ref ESGVALEPISDWDAYREELMERSGSGSKEIRQIHNRAK---RNKKRIVFAEADHLDVLKA
GOS_26940_Trans QSGVATQPIKDIDAYRDALKQTVVKSAFLMRPVFEAAS---SSARRIVFAEGEDERVLRA
gi|88706826|ref ETGVATRPVEDLVAYRERLDASVFRSSMIMRPVFAAAA---LSQRRIVFAEGEDERVLRT
gi|206686971|gb ESGVATRPIEDITAYKQKLNQTVFKSALLMRPVFEAAR---AAARRIVFAEGEDERVLRA
Spomeroyi_gi|56 ESGVARRPITDLEAYRQKLNQSVFKSALLMRPVFEAAA---KAARRLVFAEGEDERVLRA
gi|158425280|re DTGVATRPIADFDAYNEKLDEFVFRSGFIMRPLFQRAK---QDKKRVIYAEGEDERVLRA
gi|152971328|re DSGVATRPIADFDAYIEKLSEFVYKTNLFMKPIFSQAR---KEPKRVVLAEGEETRVLHA
gi|76808889|ref DGGVATRPIEDMEAYKVHLQQFVYHSGTTMKPVFQIARGAPAEKKRVVFAEGEEERVLRA
gi|187925371|re DSGVAERPIEDMEAYEQHLQQFVYHSGTTMKPIFQLARGVEPEKKRIVFAEGEEERVLRA
##################################### ################
490 500 510 520 530 540
=========+=========+=========+=========+=========+=========+
gi|86134375|ref AQRVQEEKLGLPILLGRKEVILELKEEIGFT----EDVPIFDPKTDEEKERRDRFGIAYW
GOS_26940_Trans AQAVLEETSEVPIVIGRPEVIQQRCERLGLDIRPDRDFNIVNPQQD---DRYRDYWTSYH
gi|88706826|ref AQVIVEEMTDRPILIGRPEIIARRCEKAGLTIKPGEDFEVVNPEDD---SRHRRYWEAYL
gi|206686971|gb AQAILEETTETPILIGRPEVIERRCEKLGLDVRPGRDFQLVNPEND---PRYYDYWNSYH
Spomeroyi_gi|56 AQAILEETTETPILIGRPEVIEARCEKMGLSVRPGQDFQIVNPEND---PRYYDYWTSYH
gi|158425280|re AQAVIEEGIAHPILVARPSVLEARLQRFGLSIRPGKDFEVINPEDD---PRYRDFVRSYI
gi|152971328|re TQELVSLGLAKPILVGRPSVIEMRIQKLGLQIKAGVDFEIVNNESD---PRFKEYWSEYY
gi|76808889|ref VQIVVDEKLAKPILIGRPAVIEHRIQRYGLRLTPGVDFTIVNTEHD---ERYRDFWQTYF
gi|187925371|re MQIIVDEKLAKPILIGRPAVIEQRIARYGLRLIAGQDYTVVNTDHD---ERYRDFWQEYH
############################## ########## ##########
550 560 570 580 590 600
=========+=========+=========+=========+=========+=========+
gi|86134375|ref ESRQRKGRTLTEAKKLMRERN-YFAAMMVNVGEADALITGYSRPYPTVIRPILESIQKDS
GOS_26940_Trans SLLARRGVSPDLAKSIMRTNTTAIGAVMVHRGEADSLICGAVGEFRWHLNYIEQILGSK-
gi|88706826|ref QLMSRRGVTPDLAKVIMRTNTTAIAAIMVYCGDADSMVCGSFGQYLWHLNYVRQILAYD-
gi|206686971|gb KVMQRRGVTPDLAKAIMRTNTTAIGAIMVHRGEADSLLCGTFGEYRWHLNYVQQVLGGG-
Spomeroyi_gi|56 QLMERRGVTPDIAKAIMRTNTTAIGAIMVHRGEADSLICGTFGEYRWHLNYVEQVLGSK-
gi|158425280|re EIAGRRGVTPDAARTLVRTSSTVISALAVKKGEADAMLCGIEGRFSRHLRHVRDIIGLAP
gi|152971328|re QLMKRRGITQEQAQRAVISNTTVIGAIMVHRGEADAMICGTIGEYHDHYRVVQPLFGYRD
gi|76808889|ref KMMARKGISEQLARVEMRRRTTLIGSMLVKKGEADGMICGTISTTHRHLHFIDQVIGKRA
gi|187925371|re KMMSRKGISAQMAKLEMRRRTTLIGAMLVEKGEADGMICGTVSTTHRHLHFIDQVIGKKE
##################### ##################################
610 620 630 640 650 660
=========+=========+=========+=========+=========+=========+
gi|86134375|ref GISKVAACNLMLTKQGPMFLADTTINLNPTAKDLVKISQMTSNLVKMFGMKPNVAMLSFS
GOS_26940_Trans TLSPSGALSLMILEDGPLFIADTHVWADPTPMQIAQTAKGAARHVRRFGIEPQVALCSQS
gi|88706826|ref GAHPRGALSLMITEDEPLFIADTHVHPEPTPEQIADTVMAAANHVRRFGMKPNIALCSHS
gi|206686971|gb TYSPHGALSMMILEDGPLFIADTHVHVEPTPEQIAETVIGAARHVRRFGLAPKIALCSQS
Spomeroyi_gi|56 DLRPHGALSLMILEDGPLFIADTHVRSRPSPEELAEITLGAARHVRRFGIEPQIALCSQS
gi|158425280|re GVRELAALSLLITPKGNLFLCDTQVQTEPNAADLAEMTILAAAHVRRFGIEPKVALLSHS
gi|152971328|re GVSTAGAMNALLLPSGNTFIADTYVNHDPSPEELAEITLMAAESVRRFGIEPRVALLSHS
gi|76808889|ref GCSVYGAMNALVLPGRQIFLVDTHVNVDPTPAQLAEITIMAAEEVRRFGIEPKVALLSHS
gi|187925371|re GAKVYAAMNALVLPNRQIFLVDTHVNVDPTPEQLAEITIMAAEEVRRFGIEPKIALLSHS
############################################################
670 680 690 700 710 720
=========+=========+=========+=========+=========+=========+
gi|86134375|ref NFGSTKNESSQKIREAVSYIHRNFPNAVVDGEIQADFALNPEMLAKEFPFSKLNGKKVNV
GOS_26940_Trans QFGNLNSETGKKMRQALDILDTEKVTFTYEGEMNIDTALDPELRARLLPENR--------
gi|88706826|ref QFGNLDIDSGRRVRQAMALLEAREPDFAYEGEMHIDSALDPDLRARIFPNSRLQG-PANV
gi|206686971|gb QFGNISCDTGSRLRAAIEILDDKRRDFVYEGEMNIDTALDPELRERIFPNSRLEG-AANV
Spomeroyi_gi|56 QFGNQAEGSGQRLRQAIEILDSRPRDFVYEGEMNLDSALDPELRQRIFPNSRLYG-AANV
gi|158425280|re NFGSNDTVCARRVRAALDILKDRAPELEVDGEMQAELALLPDARERILPHSRLQG-VANV
gi|152971328|re NFGSADCPSASKMRKTLELVKARAPELMIDGEMHGDAALVESIRNDRMPDSPLKG-AANI
gi|76808889|ref NFGTSNAPSAQKMRDTLAILQERAPDLHVDGEMHGDVALDAALRKEILPESTLEG-EANL
gi|187925371|re NFGTSNAPTAQKMRDTLAILRERAPDLQVDGEMHGDIALDANLRREVMPDSTLEG-DANL
###################################################
730 740 750 760 770 780
=========+=========+=========+=========+=========+=========+
gi|86134375|ref LIFPNLESANITYKLLKEMQG-AESIGPVILGLSKAVHIVQLGASVDEMVNMAALACVDA
GOS_26940_Trans ------------------------------------------------------------
gi|88706826|ref LVFAYGDAASGVRNILKMRGG-ALEVGPILMGMGNRAHIVTPSITARGLLNISALAGTDV
gi|206686971|gb LIFAHADAASGVRNILKMRAG-GLEVGPILMGMGNRAHIVSPSITARGLLNMAAIAGTPV
Spomeroyi_gi|56 LIFAHADAASGVRNVLKMKAN-GIEVGPILMGMGNRAHIVTPSITARGLLNMAAIAGTPV
gi|158425280|re LVMPDLDAADIAYNMIKVLGD-ALPVGPILMGTAKPAHILGPTVTARGIVNMTAVAVVEA
gi|152971328|re LVMPNMEAARISYNLLRVSSSEGVTVGPVLMGVAKPVHILTPIASVRRIVNMVALAVVEA
gi|76808889|ref LVLPNIDAANIAYNLLKTAAGNNIAIGPILLGAAQPVHVLTESATVRRIVNMTALLVADV
gi|187925371|re LVLPNIDAANISYNLLKTAAGNNIAIGPMLLGAAKPVHVLTASATVRRIVNMTALLVADV
======
gi|86134375|ref QQREKK
GOS_26940_Trans ------
gi|88706826|ref THYS--
gi|206686971|gb AHYG--
Spomeroyi_gi|56 AHYG--
gi|158425280|re QSEA--
gi|152971328|re QTEPL-
gi|76808889|ref NAVR--
gi|187925371|re IAAR--
Parameters used
Minimum Number Of Sequences For A Conserved Position: 5
Minimum Number Of Sequences For A Flanking Position: 8
Maximum Number Of Contiguous Nonconserved Positions: 8
Minimum Length Of A Block: 10
Allowed Gap Positions: None
Use Similarity Matrices: Yes
Flank positions of the 6 selected block(s)
Flanks: [402 457] [465 510] [517 526] [531 561] [564 597] [601 711]
New number of positions in input.fasta-gb: 288 (36% of the original 786 positions)
The phylogenetic tree in "text" format to be copied in the Annotathon "Tree" section (remember to add the taxonomic group definitions):
-------0.2-----
+------------------Congregibacter_litoralis_KT71_gi_88706826 [Add taxonomic group here!]
|
| +------Rhodobacterales_bacterium_Y4I_gi_206686971 [Add taxonomic group here!]
+---------------------+ |
| | ++
| +------++--------Silicibacter_pomeroyi_DSS-3_gi_56697770 [Add taxonomic group here!]
| |
| +-----------------GOS_26940_Translation_11-922_indirect_strand
|
+------------------+ +-----Burkholderia_pseudomallei_1710b_gi_76808889 [Add taxonomic group here!]
| | |
| | +------------------+
| | +---------+ +------Burkholderia_phytofirmans_PsJN_gi_187925371 [Add taxonomic group here!]
| | | |
| | | +------------------------Klebsiella_pneumoniae_subsp._pneumoniae_gi_152971328 [Add taxonomic group here!]
| +--+
| |
| +----------------------------Azorhizobium_caulinodans_ORS_571_gi_158425280 [Add taxonomic group here!]
|
+-----------------------------------------------------------------Polaribacter_dokdonensis_MED152_gi_86134375 [Add taxonomic group here!]